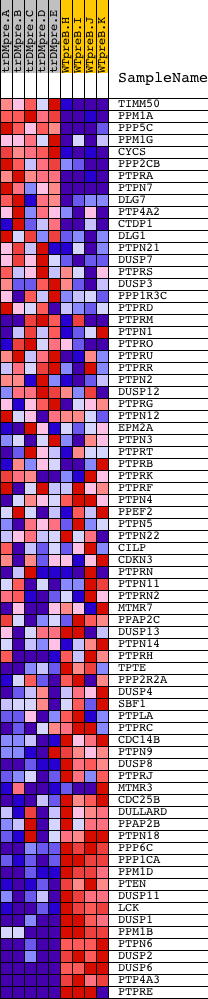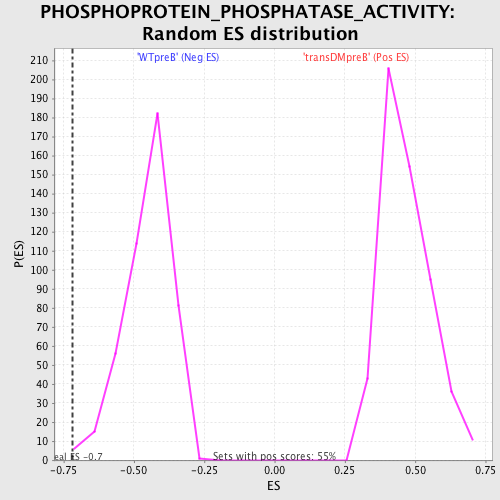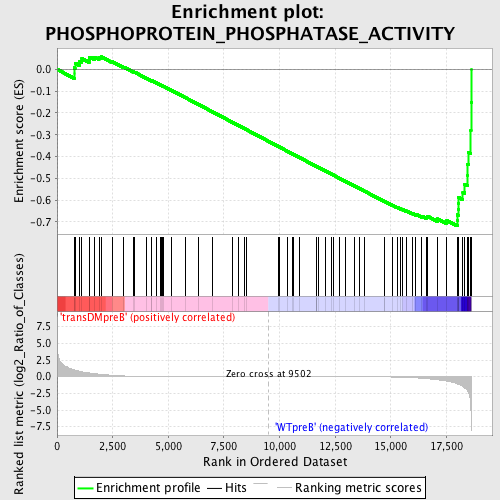
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | PHOSPHOPROTEIN_PHOSPHATASE_ACTIVITY |
| Enrichment Score (ES) | -0.7184913 |
| Normalized Enrichment Score (NES) | -1.5973114 |
| Nominal p-value | 0.0021978023 |
| FDR q-value | 0.41781902 |
| FWER p-Value | 0.513 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM50 | 17911 | 768 | 1.015 | -0.0170 | No | ||
| 2 | PPM1A | 5279 9607 | 769 | 1.015 | 0.0074 | No | ||
| 3 | PPP5C | 17952 | 808 | 0.973 | 0.0287 | No | ||
| 4 | PPM1G | 16571 | 1017 | 0.794 | 0.0366 | No | ||
| 5 | CYCS | 8821 | 1112 | 0.728 | 0.0491 | No | ||
| 6 | PPP2CB | 18636 | 1439 | 0.553 | 0.0448 | No | ||
| 7 | PTPRA | 14844 | 1463 | 0.542 | 0.0566 | No | ||
| 8 | PTPN7 | 11500 | 1675 | 0.461 | 0.0563 | No | ||
| 9 | DLG7 | 21858 | 1884 | 0.381 | 0.0542 | No | ||
| 10 | PTP4A2 | 9659 2463 2518 5324 | 1982 | 0.350 | 0.0574 | No | ||
| 11 | CTDP1 | 23399 1969 | 2488 | 0.199 | 0.0350 | No | ||
| 12 | DLG1 | 22793 1672 | 2996 | 0.108 | 0.0102 | No | ||
| 13 | PTPN21 | 21009 | 3417 | 0.065 | -0.0109 | No | ||
| 14 | DUSP7 | 10661 | 3487 | 0.060 | -0.0131 | No | ||
| 15 | PTPRS | 1507 1565 1602 22928 | 4038 | 0.037 | -0.0419 | No | ||
| 16 | DUSP3 | 1358 13022 | 4257 | 0.031 | -0.0529 | No | ||
| 17 | PPP1R3C | 23688 | 4260 | 0.031 | -0.0523 | No | ||
| 18 | PTPRD | 11554 5330 | 4447 | 0.027 | -0.0617 | No | ||
| 19 | PTPRM | 22904 | 4657 | 0.023 | -0.0724 | No | ||
| 20 | PTPN1 | 5325 | 4699 | 0.023 | -0.0741 | No | ||
| 21 | PTPRO | 1093 17256 | 4749 | 0.022 | -0.0762 | No | ||
| 22 | PTPRU | 15738 | 4781 | 0.022 | -0.0773 | No | ||
| 23 | PTPRR | 3436 19876 | 5135 | 0.017 | -0.0960 | No | ||
| 24 | PTPN2 | 23414 | 5771 | 0.012 | -0.1299 | No | ||
| 25 | DUSP12 | 13765 | 6349 | 0.009 | -0.1608 | No | ||
| 26 | PTPRG | 22097 3151 | 6990 | 0.007 | -0.1952 | No | ||
| 27 | PTPN12 | 16597 3502 3586 3665 | 7873 | 0.004 | -0.2426 | No | ||
| 28 | EPM2A | 20090 | 8152 | 0.003 | -0.2576 | No | ||
| 29 | PTPN3 | 9661 | 8442 | 0.003 | -0.2731 | No | ||
| 30 | PTPRT | 5333 | 8523 | 0.002 | -0.2773 | No | ||
| 31 | PTPRB | 19875 | 9952 | -0.001 | -0.3543 | No | ||
| 32 | PTPRK | 5332 20066 | 10001 | -0.001 | -0.3569 | No | ||
| 33 | PTPRF | 5331 2545 | 10372 | -0.002 | -0.3768 | No | ||
| 34 | PTPN4 | 13856 | 10562 | -0.003 | -0.3869 | No | ||
| 35 | PPEF2 | 16481 | 10613 | -0.003 | -0.3895 | No | ||
| 36 | PTPN5 | 17817 | 10905 | -0.004 | -0.4052 | No | ||
| 37 | PTPN22 | 1775 15470 | 11642 | -0.006 | -0.4447 | No | ||
| 38 | CILP | 19404 | 11666 | -0.006 | -0.4458 | No | ||
| 39 | CDKN3 | 16965 | 11668 | -0.006 | -0.4457 | No | ||
| 40 | PTPRN | 13914 | 11748 | -0.006 | -0.4498 | No | ||
| 41 | PTPN11 | 5326 16391 9660 | 12084 | -0.007 | -0.4677 | No | ||
| 42 | PTPRN2 | 9666 | 12316 | -0.008 | -0.4800 | No | ||
| 43 | MTMR7 | 12032 | 12400 | -0.009 | -0.4842 | No | ||
| 44 | PPAP2C | 6938 | 12681 | -0.010 | -0.4991 | No | ||
| 45 | DUSP13 | 21904 | 12976 | -0.012 | -0.5147 | No | ||
| 46 | PTPN14 | 14012 | 13380 | -0.015 | -0.5361 | No | ||
| 47 | PTPRH | 11861 | 13586 | -0.017 | -0.5467 | No | ||
| 48 | TPTE | 3875 3866 10615 3777 | 13825 | -0.019 | -0.5591 | No | ||
| 49 | PPP2R2A | 3222 21774 | 14700 | -0.036 | -0.6054 | No | ||
| 50 | DUSP4 | 18632 3820 | 14717 | -0.037 | -0.6053 | No | ||
| 51 | SBF1 | 13420 | 15066 | -0.050 | -0.6229 | No | ||
| 52 | PTPLA | 11403 6636 14680 | 15309 | -0.062 | -0.6345 | No | ||
| 53 | PTPRC | 5327 9662 | 15455 | -0.074 | -0.6405 | No | ||
| 54 | CDC14B | 3227 5750 | 15544 | -0.081 | -0.6433 | No | ||
| 55 | PTPN9 | 19434 | 15705 | -0.096 | -0.6496 | No | ||
| 56 | DUSP8 | 9493 | 15966 | -0.134 | -0.6604 | No | ||
| 57 | PTPRJ | 9664 | 16129 | -0.169 | -0.6651 | No | ||
| 58 | MTMR3 | 7908 13193 | 16383 | -0.225 | -0.6733 | No | ||
| 59 | CDC25B | 14841 | 16590 | -0.283 | -0.6776 | No | ||
| 60 | DULLARD | 20816 | 16640 | -0.296 | -0.6731 | No | ||
| 61 | PPAP2B | 7503 16161 | 17089 | -0.465 | -0.6861 | No | ||
| 62 | PTPN18 | 14279 | 17517 | -0.665 | -0.6931 | Yes | ||
| 63 | PPP6C | 7491 | 17988 | -1.061 | -0.6930 | Yes | ||
| 64 | PPP1CA | 9608 3756 914 909 | 18001 | -1.084 | -0.6676 | Yes | ||
| 65 | PPM1D | 20721 | 18030 | -1.116 | -0.6422 | Yes | ||
| 66 | PTEN | 5305 | 18041 | -1.135 | -0.6155 | Yes | ||
| 67 | DUSP11 | 17086 7785 | 18050 | -1.145 | -0.5884 | Yes | ||
| 68 | LCK | 15746 | 18240 | -1.451 | -0.5637 | Yes | ||
| 69 | DUSP1 | 23061 | 18323 | -1.672 | -0.5278 | Yes | ||
| 70 | PPM1B | 5280 1567 5281 | 18443 | -2.036 | -0.4853 | Yes | ||
| 71 | PTPN6 | 17002 | 18450 | -2.074 | -0.4357 | Yes | ||
| 72 | DUSP2 | 14865 | 18483 | -2.302 | -0.3821 | Yes | ||
| 73 | DUSP6 | 19891 3399 | 18597 | -4.571 | -0.2782 | Yes | ||
| 74 | PTP4A3 | 22460 2304 | 18605 | -5.222 | -0.1530 | Yes | ||
| 75 | PTPRE | 1662 18039 | 18614 | -6.385 | 0.0001 | Yes |