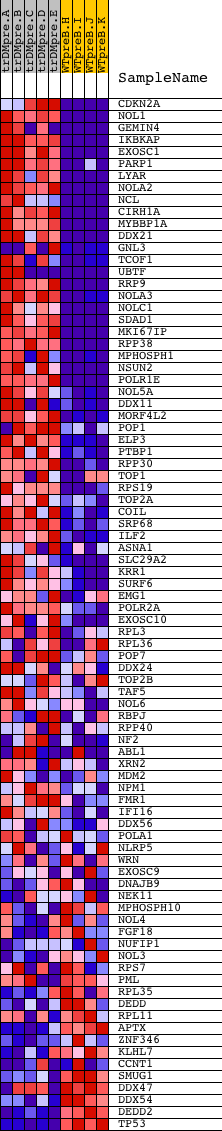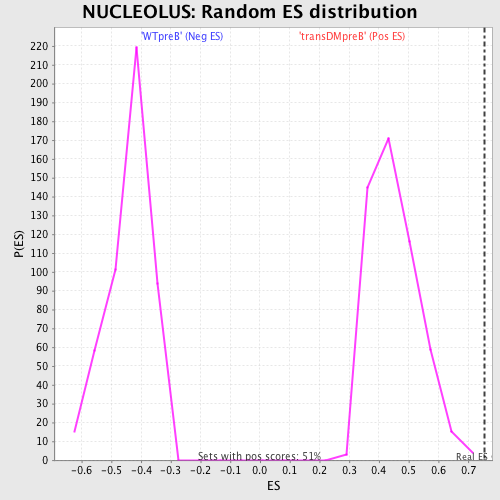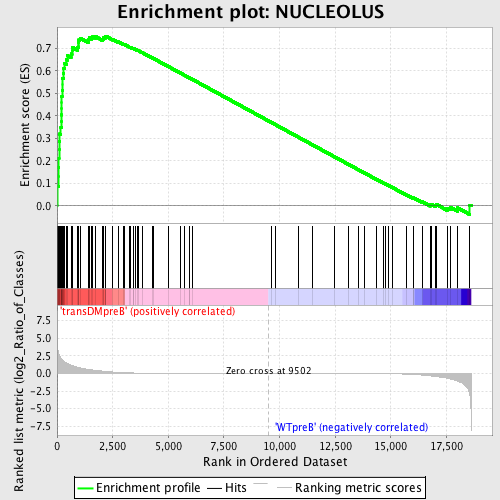
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | NUCLEOLUS |
| Enrichment Score (ES) | 0.75482893 |
| Normalized Enrichment Score (NES) | 1.6668204 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03497164 |
| FWER p-Value | 0.074 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDKN2A | 2491 15841 | 2 | 6.131 | 0.0872 | Yes | ||
| 2 | NOL1 | 8472 17280 | 52 | 3.101 | 0.1288 | Yes | ||
| 3 | GEMIN4 | 6592 6591 | 56 | 3.063 | 0.1723 | Yes | ||
| 4 | IKBKAP | 2540 6045 | 76 | 2.832 | 0.2116 | Yes | ||
| 5 | EXOSC1 | 12379 | 86 | 2.708 | 0.2497 | Yes | ||
| 6 | PARP1 | 8559 14027 | 93 | 2.632 | 0.2869 | Yes | ||
| 7 | LYAR | 16856 | 120 | 2.406 | 0.3197 | Yes | ||
| 8 | NOLA2 | 11972 | 168 | 2.153 | 0.3479 | Yes | ||
| 9 | NCL | 5153 13899 | 191 | 2.049 | 0.3759 | Yes | ||
| 10 | CIRH1A | 18478 | 198 | 2.016 | 0.4043 | Yes | ||
| 11 | MYBBP1A | 5217 1232 5216 | 206 | 1.995 | 0.4323 | Yes | ||
| 12 | DDX21 | 19751 | 209 | 1.974 | 0.4604 | Yes | ||
| 13 | GNL3 | 21894 3356 | 211 | 1.971 | 0.4884 | Yes | ||
| 14 | TCOF1 | 5660 | 225 | 1.917 | 0.5150 | Yes | ||
| 15 | UBTF | 1313 10052 | 236 | 1.867 | 0.5411 | Yes | ||
| 16 | RRP9 | 19328 | 237 | 1.865 | 0.5676 | Yes | ||
| 17 | NOLA3 | 14914 | 280 | 1.717 | 0.5898 | Yes | ||
| 18 | NOLC1 | 7704 | 300 | 1.658 | 0.6124 | Yes | ||
| 19 | SDAD1 | 16479 | 333 | 1.600 | 0.6335 | Yes | ||
| 20 | MKI67IP | 7509 | 429 | 1.469 | 0.6493 | Yes | ||
| 21 | RPP38 | 14698 | 452 | 1.436 | 0.6686 | Yes | ||
| 22 | MPHOSPH1 | 23878 | 629 | 1.163 | 0.6757 | Yes | ||
| 23 | NSUN2 | 21610 | 671 | 1.108 | 0.6892 | Yes | ||
| 24 | POLR1E | 7226 | 677 | 1.101 | 0.7047 | Yes | ||
| 25 | NOL5A | 12474 | 928 | 0.871 | 0.7036 | Yes | ||
| 26 | DDX11 | 23169 | 951 | 0.849 | 0.7145 | Yes | ||
| 27 | MORF4L2 | 12118 | 953 | 0.844 | 0.7264 | Yes | ||
| 28 | POP1 | 2214 7482 | 967 | 0.837 | 0.7377 | Yes | ||
| 29 | ELP3 | 21782 | 1028 | 0.786 | 0.7456 | Yes | ||
| 30 | PTBP1 | 5303 | 1403 | 0.569 | 0.7335 | Yes | ||
| 31 | RPP30 | 23877 | 1425 | 0.558 | 0.7404 | Yes | ||
| 32 | TOP1 | 5790 5789 10210 | 1435 | 0.556 | 0.7478 | Yes | ||
| 33 | RPS19 | 5398 | 1565 | 0.505 | 0.7480 | Yes | ||
| 34 | TOP2A | 20257 | 1604 | 0.489 | 0.7529 | Yes | ||
| 35 | COIL | 20708 | 1736 | 0.442 | 0.7522 | Yes | ||
| 36 | SRP68 | 20143 | 2035 | 0.331 | 0.7408 | Yes | ||
| 37 | ILF2 | 12582 | 2071 | 0.321 | 0.7435 | Yes | ||
| 38 | ASNA1 | 18810 | 2077 | 0.320 | 0.7478 | Yes | ||
| 39 | SLC29A2 | 23775 | 2158 | 0.295 | 0.7477 | Yes | ||
| 40 | KRR1 | 6989 3313 | 2166 | 0.293 | 0.7515 | Yes | ||
| 41 | SURF6 | 9945 | 2181 | 0.288 | 0.7548 | Yes | ||
| 42 | EMG1 | 1016 17005 | 2506 | 0.196 | 0.7401 | No | ||
| 43 | POLR2A | 5394 | 2747 | 0.149 | 0.7293 | No | ||
| 44 | EXOSC10 | 2482 11943 | 2977 | 0.109 | 0.7185 | No | ||
| 45 | RPL3 | 11330 | 3017 | 0.105 | 0.7179 | No | ||
| 46 | RPL36 | 12024 | 3261 | 0.077 | 0.7059 | No | ||
| 47 | POP7 | 16329 | 3298 | 0.074 | 0.7050 | No | ||
| 48 | DDX24 | 2084 20995 | 3424 | 0.064 | 0.6992 | No | ||
| 49 | TOP2B | 5791 | 3451 | 0.063 | 0.6987 | No | ||
| 50 | TAF5 | 23833 5934 3759 | 3501 | 0.059 | 0.6969 | No | ||
| 51 | NOL6 | 2385 2405 15913 2466 | 3596 | 0.054 | 0.6926 | No | ||
| 52 | RBPJ | 9709 3594 5370 | 3651 | 0.052 | 0.6904 | No | ||
| 53 | RPP40 | 21491 | 3829 | 0.044 | 0.6815 | No | ||
| 54 | NF2 | 1222 5166 | 4275 | 0.030 | 0.6579 | No | ||
| 55 | ABL1 | 2693 4301 2794 | 4331 | 0.029 | 0.6553 | No | ||
| 56 | XRN2 | 2936 6302 2905 | 4992 | 0.019 | 0.6200 | No | ||
| 57 | MDM2 | 19620 3327 | 5559 | 0.014 | 0.5896 | No | ||
| 58 | NPM1 | 1196 | 5715 | 0.012 | 0.5814 | No | ||
| 59 | FMR1 | 24321 | 5960 | 0.011 | 0.5684 | No | ||
| 60 | IFI16 | 13745 | 5962 | 0.011 | 0.5685 | No | ||
| 61 | DDX56 | 6984 | 6107 | 0.010 | 0.5609 | No | ||
| 62 | POLA1 | 24112 | 9653 | -0.000 | 0.3696 | No | ||
| 63 | NLRP5 | 10743 18358 | 9798 | -0.001 | 0.3619 | No | ||
| 64 | WRN | 5882 10315 | 10865 | -0.003 | 0.3044 | No | ||
| 65 | EXOSC9 | 15610 | 11477 | -0.005 | 0.2715 | No | ||
| 66 | DNAJB9 | 6565 | 12447 | -0.009 | 0.2193 | No | ||
| 67 | NEK11 | 3099 19016 | 13095 | -0.013 | 0.1846 | No | ||
| 68 | MPHOSPH10 | 17805 | 13549 | -0.016 | 0.1604 | No | ||
| 69 | NOL4 | 2028 11432 | 13819 | -0.019 | 0.1461 | No | ||
| 70 | FGF18 | 4721 1351 | 14368 | -0.028 | 0.1169 | No | ||
| 71 | NUFIP1 | 21955 | 14685 | -0.036 | 0.1004 | No | ||
| 72 | NOL3 | 18496 915 921 | 14751 | -0.038 | 0.0974 | No | ||
| 73 | RPS7 | 9760 | 14888 | -0.043 | 0.0907 | No | ||
| 74 | PML | 3015 3074 3020 5270 | 15074 | -0.050 | 0.0814 | No | ||
| 75 | RPL35 | 12360 | 15718 | -0.097 | 0.0481 | No | ||
| 76 | DEDD | 14056 | 16010 | -0.142 | 0.0344 | No | ||
| 77 | RPL11 | 12450 | 16025 | -0.143 | 0.0357 | No | ||
| 78 | APTX | 2455 7291 | 16402 | -0.230 | 0.0187 | No | ||
| 79 | ZNF346 | 3163 6517 3193 | 16763 | -0.338 | 0.0041 | No | ||
| 80 | KLHL7 | 16907 | 16815 | -0.357 | 0.0064 | No | ||
| 81 | CCNT1 | 22140 11607 | 17006 | -0.435 | 0.0024 | No | ||
| 82 | SMUG1 | 22109 | 17033 | -0.443 | 0.0073 | No | ||
| 83 | DDX47 | 17261 12577 1133 | 17533 | -0.678 | -0.0100 | No | ||
| 84 | DDX54 | 7778 | 17683 | -0.778 | -0.0069 | No | ||
| 85 | DEDD2 | 7429 | 17995 | -1.072 | -0.0084 | No | ||
| 86 | TP53 | 20822 | 18549 | -2.941 | 0.0036 | No |