Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
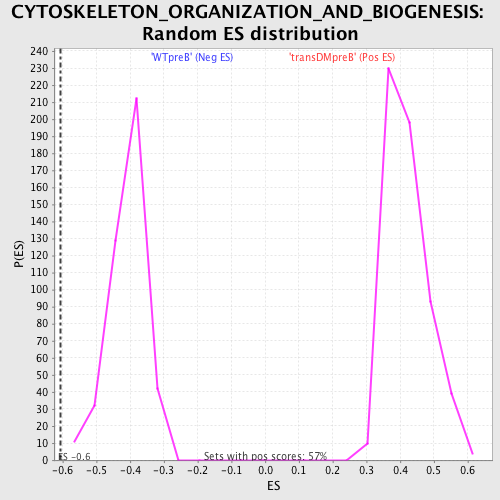
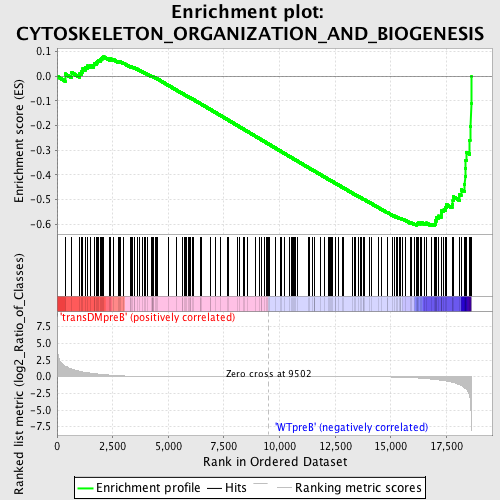
| GeneSet | CYTOSKELETON_ORGANIZATION_AND_BIOGENESIS |
| Enrichment Score (ES) | -0.6073559 |
| Normalized Enrichment Score (NES) | -1.488881 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.38826728 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ANLN | 7581 3138 12730 | 363 | 1.556 | 0.0097 | No | ||
| 2 | RCC1 | 15735 | 655 | 1.132 | 0.0153 | No | ||
| 3 | TUBG1 | 20662 | 1021 | 0.790 | 0.0104 | No | ||
| 4 | KIF23 | 19091 | 1107 | 0.731 | 0.0195 | No | ||
| 5 | ATP2C1 | 10660 6178 | 1157 | 0.698 | 0.0301 | No | ||
| 6 | TUBGCP2 | 17576 | 1259 | 0.639 | 0.0366 | No | ||
| 7 | KPNA2 | 9245 | 1363 | 0.588 | 0.0422 | No | ||
| 8 | NUBP1 | 11171 22860 | 1507 | 0.526 | 0.0443 | No | ||
| 9 | HOOK3 | 18903 6734 | 1658 | 0.468 | 0.0450 | No | ||
| 10 | FGD6 | 19901 | 1659 | 0.467 | 0.0538 | No | ||
| 11 | SOD1 | 9846 | 1769 | 0.431 | 0.0560 | No | ||
| 12 | KPTN | 18385 | 1809 | 0.411 | 0.0617 | No | ||
| 13 | MYH9 | 2252 2244 | 1871 | 0.386 | 0.0657 | No | ||
| 14 | CDC42BPB | 20982 | 1959 | 0.357 | 0.0677 | No | ||
| 15 | KIF4A | 9221 24281 | 1988 | 0.348 | 0.0727 | No | ||
| 16 | FLNB | 13742 | 2037 | 0.331 | 0.0764 | No | ||
| 17 | STMN1 | 9261 | 2099 | 0.313 | 0.0790 | No | ||
| 18 | MAP1S | 11279 | 2376 | 0.233 | 0.0684 | No | ||
| 19 | CFL1 | 4516 | 2394 | 0.228 | 0.0718 | No | ||
| 20 | LIMK1 | 16350 | 2527 | 0.190 | 0.0682 | No | ||
| 21 | DOCK2 | 8243 | 2758 | 0.147 | 0.0585 | No | ||
| 22 | RHOT1 | 12227 | 2784 | 0.141 | 0.0598 | No | ||
| 23 | THY1 | 19481 | 2833 | 0.132 | 0.0597 | No | ||
| 24 | DLG1 | 22793 1672 | 2996 | 0.108 | 0.0530 | No | ||
| 25 | CAPG | 17411 | 3296 | 0.074 | 0.0382 | No | ||
| 26 | AP1M2 | 19209 8600 4393 | 3308 | 0.073 | 0.0389 | No | ||
| 27 | MAPT | 9420 1347 | 3342 | 0.071 | 0.0385 | No | ||
| 28 | TTK | 19366 | 3392 | 0.067 | 0.0371 | No | ||
| 29 | RHOJ | 21241 | 3468 | 0.061 | 0.0342 | No | ||
| 30 | FSCN1 | 16637 | 3469 | 0.061 | 0.0353 | No | ||
| 31 | FLNA | 24129 | 3592 | 0.054 | 0.0297 | No | ||
| 32 | APOE | 8618 | 3698 | 0.049 | 0.0250 | No | ||
| 33 | GHSR | 1810 15625 | 3850 | 0.043 | 0.0176 | No | ||
| 34 | EVL | 21156 | 3935 | 0.040 | 0.0138 | No | ||
| 35 | ARPC1A | 3541 16628 3519 | 3988 | 0.038 | 0.0117 | No | ||
| 36 | WASF1 | 13514 | 4070 | 0.036 | 0.0080 | No | ||
| 37 | PLEK2 | 21033 | 4234 | 0.031 | -0.0003 | No | ||
| 38 | BCAR1 | 18741 | 4267 | 0.031 | -0.0014 | No | ||
| 39 | TSC1 | 7235 | 4273 | 0.030 | -0.0011 | No | ||
| 40 | NF2 | 1222 5166 | 4275 | 0.030 | -0.0006 | No | ||
| 41 | ABL1 | 2693 4301 2794 | 4331 | 0.029 | -0.0030 | No | ||
| 42 | PAK1 | 9527 | 4349 | 0.029 | -0.0034 | No | ||
| 43 | ANK3 | 8591 3445 3304 3381 | 4426 | 0.027 | -0.0070 | No | ||
| 44 | FGD4 | 10306 5870 1717 1669 1700 | 4454 | 0.027 | -0.0080 | No | ||
| 45 | MID1 | 5097 5098 | 4528 | 0.025 | -0.0115 | No | ||
| 46 | RHOF | 16379 | 5016 | 0.019 | -0.0375 | No | ||
| 47 | ARFIP2 | 1226 17702 | 5367 | 0.015 | -0.0562 | No | ||
| 48 | RND3 | 13181 | 5624 | 0.013 | -0.0698 | No | ||
| 49 | NEBL | 14678 2691 | 5648 | 0.013 | -0.0708 | No | ||
| 50 | MYH6 | 9436 | 5731 | 0.012 | -0.0750 | No | ||
| 51 | CCL3 | 20317 | 5756 | 0.012 | -0.0761 | No | ||
| 52 | NEFL | 9459 | 5801 | 0.012 | -0.0783 | No | ||
| 53 | DYNC1I1 | 17528 | 5890 | 0.011 | -0.0828 | No | ||
| 54 | TTN | 14552 2743 14553 | 5958 | 0.011 | -0.0863 | No | ||
| 55 | MYOZ1 | 21907 | 5978 | 0.011 | -0.0871 | No | ||
| 56 | NCK1 | 9447 5152 | 6001 | 0.011 | -0.0881 | No | ||
| 57 | RAC3 | 20561 | 6105 | 0.010 | -0.0935 | No | ||
| 58 | PRKG1 | 5289 | 6119 | 0.010 | -0.0940 | No | ||
| 59 | KISS1 | 6603 | 6435 | 0.009 | -0.1109 | No | ||
| 60 | KIF1A | 9216 | 6456 | 0.009 | -0.1118 | No | ||
| 61 | RHOA | 8624 4409 4410 | 6499 | 0.009 | -0.1139 | No | ||
| 62 | CXCL12 | 9792 1182 1031 | 6882 | 0.007 | -0.1345 | No | ||
| 63 | NLGN1 | 15368 | 6911 | 0.007 | -0.1359 | No | ||
| 64 | MRAS | 9409 | 7136 | 0.006 | -0.1479 | No | ||
| 65 | CDC42 | 4503 8722 4504 2465 | 7364 | 0.005 | -0.1601 | No | ||
| 66 | MYH10 | 8077 | 7678 | 0.005 | -0.1770 | No | ||
| 67 | MAST1 | 12143 | 7711 | 0.005 | -0.1786 | No | ||
| 68 | SSH1 | 10540 | 8097 | 0.004 | -0.1994 | No | ||
| 69 | APBA1 | 11483 | 8189 | 0.003 | -0.2043 | No | ||
| 70 | CDC42EP5 | 17991 | 8366 | 0.003 | -0.2138 | No | ||
| 71 | ACTA1 | 18715 | 8440 | 0.003 | -0.2177 | No | ||
| 72 | CLASP1 | 14159 | 8552 | 0.002 | -0.2237 | No | ||
| 73 | CDC42BPA | 10384 | 8920 | 0.001 | -0.2436 | No | ||
| 74 | DNAJB6 | 6246 3538 | 9087 | 0.001 | -0.2525 | No | ||
| 75 | TESK2 | 10504 | 9170 | 0.001 | -0.2570 | No | ||
| 76 | PLD2 | 20803 | 9178 | 0.001 | -0.2573 | No | ||
| 77 | FGD1 | 24228 | 9342 | 0.000 | -0.2662 | No | ||
| 78 | SNAP23 | 2853 14893 | 9419 | 0.000 | -0.2703 | No | ||
| 79 | SEPT5 | 9598 | 9438 | 0.000 | -0.2712 | No | ||
| 80 | PYY | 20207 | 9515 | -0.000 | -0.2754 | No | ||
| 81 | KIFAP3 | 3962 14078 9224 | 9555 | -0.000 | -0.2775 | No | ||
| 82 | CLASP2 | 13338 | 9834 | -0.001 | -0.2925 | No | ||
| 83 | PACSIN2 | 2312 | 10040 | -0.001 | -0.3036 | No | ||
| 84 | OPA1 | 13169 22800 7894 | 10063 | -0.001 | -0.3048 | No | ||
| 85 | ADRA2A | 4355 | 10205 | -0.002 | -0.3124 | No | ||
| 86 | MTSS1 | 5585 22285 | 10467 | -0.002 | -0.3265 | No | ||
| 87 | KIF5B | 4955 2031 | 10526 | -0.003 | -0.3296 | No | ||
| 88 | NEXN | 12737 1903 | 10587 | -0.003 | -0.3328 | No | ||
| 89 | DSTN | 14823 | 10605 | -0.003 | -0.3337 | No | ||
| 90 | TAOK2 | 10610 3794 | 10674 | -0.003 | -0.3373 | No | ||
| 91 | KIF5A | 9222 | 10720 | -0.003 | -0.3397 | No | ||
| 92 | CXCL1 | 9044 | 10817 | -0.003 | -0.3448 | No | ||
| 93 | MYH11 | 22656 | 11302 | -0.005 | -0.3710 | No | ||
| 94 | TUBGCP5 | 6123 2197 | 11351 | -0.005 | -0.3735 | No | ||
| 95 | FSCN2 | 20569 | 11496 | -0.005 | -0.3812 | No | ||
| 96 | KIF11 | 23873 | 11575 | -0.006 | -0.3853 | No | ||
| 97 | MARK4 | 10572 | 11576 | -0.006 | -0.3852 | No | ||
| 98 | ARHGEF17 | 17737 | 11587 | -0.006 | -0.3857 | No | ||
| 99 | ABI2 | 4037 6830 | 11849 | -0.006 | -0.3997 | No | ||
| 100 | GHRL | 1183 17040 | 12030 | -0.007 | -0.4093 | No | ||
| 101 | DBN1 | 21452 | 12198 | -0.008 | -0.4182 | No | ||
| 102 | SORBS3 | 21766 | 12235 | -0.008 | -0.4200 | No | ||
| 103 | UXT | 5837 10267 | 12291 | -0.008 | -0.4228 | No | ||
| 104 | VIL1 | 14224 | 12320 | -0.008 | -0.4242 | No | ||
| 105 | TNXB | 1557 8174 878 8175 | 12365 | -0.009 | -0.4264 | No | ||
| 106 | MARK1 | 5955 | 12495 | -0.009 | -0.4333 | No | ||
| 107 | KRT20 | 12410 | 12514 | -0.009 | -0.4341 | No | ||
| 108 | DES | 14215 | 12630 | -0.010 | -0.4401 | No | ||
| 109 | ARHGEF2 | 4987 | 12818 | -0.011 | -0.4500 | No | ||
| 110 | PAFAH1B1 | 1340 5220 9524 | 12851 | -0.011 | -0.4516 | No | ||
| 111 | PCLO | 11207 6495 | 13266 | -0.014 | -0.4737 | No | ||
| 112 | PLA2G1B | 9581 | 13366 | -0.015 | -0.4788 | No | ||
| 113 | SMC3 | 8811 23815 8812 4571 | 13400 | -0.015 | -0.4804 | No | ||
| 114 | SPTBN4 | 13471 1104 | 13558 | -0.016 | -0.4886 | No | ||
| 115 | FGD5 | 10555 17352 | 13614 | -0.017 | -0.4912 | No | ||
| 116 | AMOT | 11341 24037 6587 | 13661 | -0.017 | -0.4934 | No | ||
| 117 | CRK | 4559 1249 | 13772 | -0.019 | -0.4990 | No | ||
| 118 | DST | 3934 3984 4644 3976 14281 4070 4020 4008 4040 | 13776 | -0.019 | -0.4988 | No | ||
| 119 | WASF3 | 16625 | 13817 | -0.019 | -0.5006 | No | ||
| 120 | CDC42EP2 | 4180 14771 | 14041 | -0.022 | -0.5123 | No | ||
| 121 | KIF2C | 15789 | 14133 | -0.024 | -0.5168 | No | ||
| 122 | RND1 | 5868 | 14441 | -0.029 | -0.5329 | No | ||
| 123 | PSCD2 | 17826 | 14566 | -0.032 | -0.5390 | No | ||
| 124 | ARHGAP10 | 3808 18836 | 14850 | -0.041 | -0.5535 | No | ||
| 125 | DMD | 24295 2647 | 14851 | -0.041 | -0.5528 | No | ||
| 126 | MYH7 | 21828 4709 | 15059 | -0.050 | -0.5631 | No | ||
| 127 | RACGAP1 | 2240 22132 | 15145 | -0.053 | -0.5667 | No | ||
| 128 | KIF13B | 21982 21983 | 15239 | -0.058 | -0.5706 | No | ||
| 129 | APC | 4396 2022 | 15290 | -0.061 | -0.5722 | No | ||
| 130 | ROCK1 | 5386 | 15375 | -0.067 | -0.5754 | No | ||
| 131 | NF1 | 5165 | 15418 | -0.071 | -0.5764 | No | ||
| 132 | GSN | 2784 | 15449 | -0.073 | -0.5766 | No | ||
| 133 | KIF1B | 15671 2476 4950 15673 | 15536 | -0.081 | -0.5798 | No | ||
| 134 | CENTD2 | 1758 18169 2284 | 15643 | -0.090 | -0.5838 | No | ||
| 135 | DYNLL1 | 12135 | 15881 | -0.117 | -0.5944 | No | ||
| 136 | LRPPRC | 13031 22883 | 15937 | -0.128 | -0.5950 | No | ||
| 137 | ARHGDIB | 16950 | 16075 | -0.157 | -0.5995 | No | ||
| 138 | MYO1E | 7742 | 16157 | -0.173 | -0.6006 | Yes | ||
| 139 | MID1IP1 | 24381 | 16182 | -0.181 | -0.5985 | Yes | ||
| 140 | RHOT2 | 1533 10073 | 16201 | -0.185 | -0.5960 | Yes | ||
| 141 | RASA1 | 10174 | 16249 | -0.195 | -0.5948 | Yes | ||
| 142 | MYO9B | 5142 18581 | 16339 | -0.214 | -0.5956 | Yes | ||
| 143 | MYO6 | 19368 | 16356 | -0.219 | -0.5924 | Yes | ||
| 144 | LRMP | 17244 | 16509 | -0.261 | -0.5957 | Yes | ||
| 145 | NCK2 | 9448 | 16599 | -0.287 | -0.5951 | Yes | ||
| 146 | MAPRE1 | 4652 | 16819 | -0.360 | -0.6002 | Yes | ||
| 147 | MYL6 | 9438 3408 | 16952 | -0.412 | -0.5996 | Yes | ||
| 148 | TNNT2 | 14113 | 17013 | -0.437 | -0.5946 | Yes | ||
| 149 | ARF6 | 21252 | 17016 | -0.438 | -0.5864 | Yes | ||
| 150 | SSH2 | 6202 | 17036 | -0.443 | -0.5791 | Yes | ||
| 151 | RAC1 | 16302 | 17052 | -0.451 | -0.5714 | Yes | ||
| 152 | PRC1 | 12862 18203 | 17136 | -0.482 | -0.5668 | Yes | ||
| 153 | FGD2 | 23309 | 17261 | -0.528 | -0.5636 | Yes | ||
| 154 | ARPC4 | 12642 | 17266 | -0.530 | -0.5538 | Yes | ||
| 155 | WASL | 17203 | 17292 | -0.543 | -0.5449 | Yes | ||
| 156 | FGD3 | 11399 | 17389 | -0.590 | -0.5390 | Yes | ||
| 157 | STX5 | 23942 | 17470 | -0.636 | -0.5313 | Yes | ||
| 158 | MAP3K11 | 11163 | 17499 | -0.655 | -0.5205 | Yes | ||
| 159 | CENPJ | 21809 23694 | 17754 | -0.831 | -0.5186 | Yes | ||
| 160 | WIPF1 | 14560 | 17791 | -0.863 | -0.5042 | Yes | ||
| 161 | ARHGAP4 | 9343 | 17817 | -0.882 | -0.4890 | Yes | ||
| 162 | PDPK1 | 23097 | 18082 | -1.189 | -0.4808 | Yes | ||
| 163 | SCIN | 21079 | 18160 | -1.285 | -0.4608 | Yes | ||
| 164 | WASF2 | 6326 | 18318 | -1.668 | -0.4378 | Yes | ||
| 165 | SNAP29 | 22837 | 18334 | -1.700 | -0.4065 | Yes | ||
| 166 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 18347 | -1.737 | -0.3744 | Yes | ||
| 167 | SMC1A | 2572 24225 6279 10780 | 18370 | -1.797 | -0.3417 | Yes | ||
| 168 | CNN2 | 19953 3391 | 18391 | -1.835 | -0.3082 | Yes | ||
| 169 | LASP1 | 4986 4985 1248 | 18546 | -2.918 | -0.2615 | Yes | ||
| 170 | ARPC5 | 12580 7487 | 18561 | -3.137 | -0.2031 | Yes | ||
| 171 | NUSAP1 | 2956 4250 | 18603 | -5.121 | -0.1087 | Yes | ||
| 172 | RAN | 5356 9691 | 18611 | -5.795 | 0.0003 | Yes |