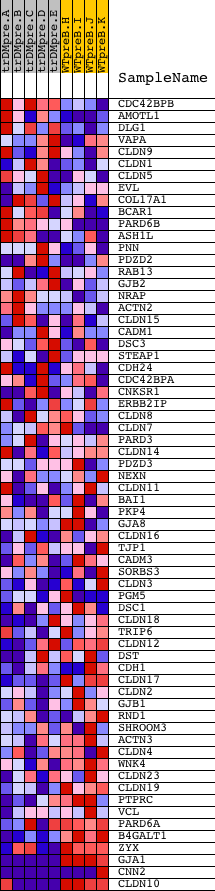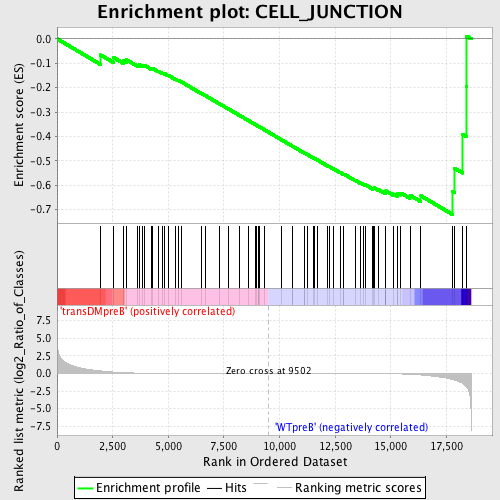
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | CELL_JUNCTION |
| Enrichment Score (ES) | -0.71964514 |
| Normalized Enrichment Score (NES) | -1.5443592 |
| Nominal p-value | 0.00625 |
| FDR q-value | 0.3658562 |
| FWER p-Value | 0.886 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDC42BPB | 20982 | 1959 | 0.357 | -0.0661 | No | ||
| 2 | AMOTL1 | 19242 | 2546 | 0.186 | -0.0770 | No | ||
| 3 | DLG1 | 22793 1672 | 2996 | 0.108 | -0.0893 | No | ||
| 4 | VAPA | 22908 | 3125 | 0.091 | -0.0862 | No | ||
| 5 | CLDN9 | 24470 | 3629 | 0.052 | -0.1075 | No | ||
| 6 | CLDN1 | 22623 | 3712 | 0.049 | -0.1065 | No | ||
| 7 | CLDN5 | 22826 | 3818 | 0.044 | -0.1072 | No | ||
| 8 | EVL | 21156 | 3935 | 0.040 | -0.1090 | No | ||
| 9 | COL17A1 | 23647 3671 | 4224 | 0.031 | -0.1211 | No | ||
| 10 | BCAR1 | 18741 | 4267 | 0.031 | -0.1200 | No | ||
| 11 | PARD6B | 12209 | 4546 | 0.025 | -0.1322 | No | ||
| 12 | ASH1L | 5311 15546 15547 | 4752 | 0.022 | -0.1408 | No | ||
| 13 | PNN | 9596 | 4829 | 0.021 | -0.1426 | No | ||
| 14 | PDZD2 | 22321 2174 22323 22322 | 5008 | 0.019 | -0.1501 | No | ||
| 15 | RAB13 | 15535 | 5315 | 0.016 | -0.1649 | No | ||
| 16 | GJB2 | 9018 21806 | 5441 | 0.015 | -0.1700 | No | ||
| 17 | NRAP | 23642 | 5571 | 0.013 | -0.1755 | No | ||
| 18 | ACTN2 | 21545 | 6480 | 0.009 | -0.2235 | No | ||
| 19 | CLDN15 | 16667 3572 | 6679 | 0.008 | -0.2333 | No | ||
| 20 | CADM1 | 7057 | 7306 | 0.006 | -0.2664 | No | ||
| 21 | DSC3 | 23487 | 7705 | 0.005 | -0.2873 | No | ||
| 22 | STEAP1 | 16926 | 8188 | 0.003 | -0.3129 | No | ||
| 23 | CDH24 | 10726 | 8602 | 0.002 | -0.3350 | No | ||
| 24 | CDC42BPA | 10384 | 8920 | 0.001 | -0.3519 | No | ||
| 25 | CNKSR1 | 15719 20331 | 8947 | 0.001 | -0.3531 | No | ||
| 26 | ERBB2IP | 3232 12234 | 9066 | 0.001 | -0.3594 | No | ||
| 27 | CLDN8 | 22549 1747 | 9068 | 0.001 | -0.3593 | No | ||
| 28 | CLDN7 | 20817 | 9090 | 0.001 | -0.3603 | No | ||
| 29 | PARD3 | 8212 13553 | 9339 | 0.000 | -0.3737 | No | ||
| 30 | CLDN14 | 12081 | 10094 | -0.002 | -0.4141 | No | ||
| 31 | PDZD3 | 19153 | 10568 | -0.003 | -0.4393 | No | ||
| 32 | NEXN | 12737 1903 | 10587 | -0.003 | -0.4400 | No | ||
| 33 | CLDN11 | 15616 | 11118 | -0.004 | -0.4681 | No | ||
| 34 | BAI1 | 22459 | 11136 | -0.004 | -0.4686 | No | ||
| 35 | PKP4 | 2822 15008 | 11243 | -0.004 | -0.4738 | No | ||
| 36 | GJA8 | 15234 | 11543 | -0.005 | -0.4893 | No | ||
| 37 | CLDN16 | 22802 | 11567 | -0.005 | -0.4899 | No | ||
| 38 | TJP1 | 17803 | 11707 | -0.006 | -0.4968 | No | ||
| 39 | CADM3 | 13746 | 12169 | -0.008 | -0.5208 | No | ||
| 40 | SORBS3 | 21766 | 12235 | -0.008 | -0.5234 | No | ||
| 41 | CLDN3 | 16680 | 12420 | -0.009 | -0.5323 | No | ||
| 42 | PGM5 | 5928 | 12730 | -0.010 | -0.5478 | No | ||
| 43 | DSC1 | 8864 | 12882 | -0.011 | -0.5548 | No | ||
| 44 | CLDN18 | 19028 | 12890 | -0.011 | -0.5539 | No | ||
| 45 | TRIP6 | 16331 | 13397 | -0.015 | -0.5795 | No | ||
| 46 | CLDN12 | 16925 | 13636 | -0.017 | -0.5905 | No | ||
| 47 | DST | 3934 3984 4644 3976 14281 4070 4020 4008 4040 | 13776 | -0.019 | -0.5959 | No | ||
| 48 | CDH1 | 18479 | 13858 | -0.020 | -0.5981 | No | ||
| 49 | CLDN17 | 10757 | 14197 | -0.025 | -0.6136 | No | ||
| 50 | CLDN2 | 4527 | 14207 | -0.025 | -0.6113 | No | ||
| 51 | GJB1 | 24276 | 14245 | -0.026 | -0.6104 | No | ||
| 52 | RND1 | 5868 | 14441 | -0.029 | -0.6177 | No | ||
| 53 | SHROOM3 | 428 | 14753 | -0.038 | -0.6302 | No | ||
| 54 | ACTN3 | 23967 8540 | 14766 | -0.038 | -0.6267 | No | ||
| 55 | CLDN4 | 8747 | 14781 | -0.039 | -0.6231 | No | ||
| 56 | WNK4 | 7644 | 15120 | -0.052 | -0.6356 | No | ||
| 57 | CLDN23 | 18893 | 15282 | -0.061 | -0.6376 | No | ||
| 58 | CLDN19 | 16105 472 | 15315 | -0.062 | -0.6324 | No | ||
| 59 | PTPRC | 5327 9662 | 15455 | -0.074 | -0.6317 | No | ||
| 60 | VCL | 22083 | 15862 | -0.115 | -0.6409 | No | ||
| 61 | PARD6A | 12142 | 16353 | -0.219 | -0.6431 | Yes | ||
| 62 | B4GALT1 | 15918 | 17774 | -0.844 | -0.6261 | Yes | ||
| 63 | ZYX | 10443 | 17858 | -0.912 | -0.5296 | Yes | ||
| 64 | GJA1 | 20023 | 18234 | -1.435 | -0.3909 | Yes | ||
| 65 | CNN2 | 19953 3391 | 18391 | -1.835 | -0.1962 | Yes | ||
| 66 | CLDN10 | 21935 2785 2601 2720 7188 | 18410 | -1.880 | 0.0111 | Yes |