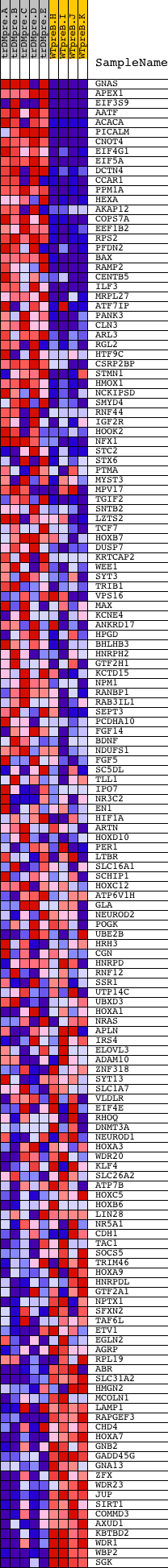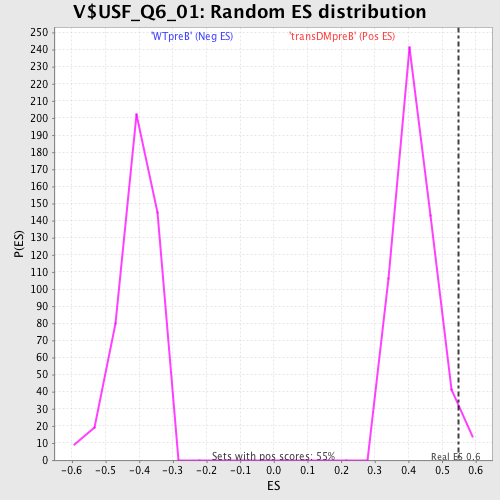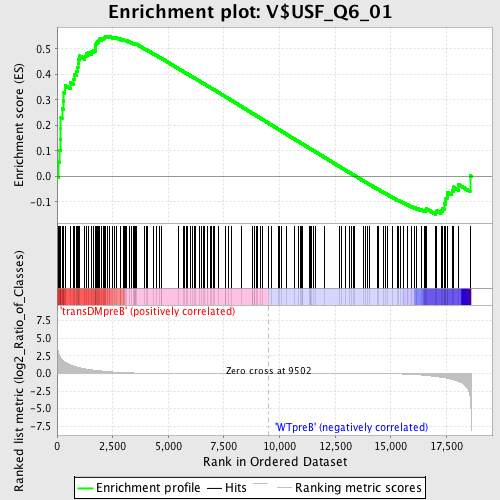
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | V$USF_Q6_01 |
| Enrichment Score (ES) | 0.55027694 |
| Normalized Enrichment Score (NES) | 1.3069502 |
| Nominal p-value | 0.033027522 |
| FDR q-value | 0.5034579 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GNAS | 9025 2963 2752 | 54 | 3.075 | 0.0583 | Yes | ||
| 2 | APEX1 | 22028 | 117 | 2.422 | 0.1032 | Yes | ||
| 3 | EIF3S9 | 16643 3528 | 158 | 2.211 | 0.1451 | Yes | ||
| 4 | AATF | 20315 | 170 | 2.146 | 0.1872 | Yes | ||
| 5 | ACACA | 309 4209 | 172 | 2.135 | 0.2297 | Yes | ||
| 6 | PICALM | 18191 | 223 | 1.919 | 0.2652 | Yes | ||
| 7 | CNOT4 | 7010 1022 | 276 | 1.731 | 0.2968 | Yes | ||
| 8 | EIF4G1 | 22818 | 305 | 1.644 | 0.3281 | Yes | ||
| 9 | EIF5A | 11345 20379 6590 | 354 | 1.574 | 0.3568 | Yes | ||
| 10 | DCTN4 | 12560 7474 | 589 | 1.207 | 0.3682 | Yes | ||
| 11 | CCAR1 | 7454 | 748 | 1.030 | 0.3801 | Yes | ||
| 12 | PPM1A | 5279 9607 | 769 | 1.015 | 0.3993 | Yes | ||
| 13 | HEXA | 19420 | 854 | 0.930 | 0.4132 | Yes | ||
| 14 | AKAP12 | 17600 | 903 | 0.890 | 0.4284 | Yes | ||
| 15 | COPS7A | 11221 16997 | 941 | 0.857 | 0.4434 | Yes | ||
| 16 | EEF1B2 | 4131 12063 | 962 | 0.839 | 0.4591 | Yes | ||
| 17 | RPS2 | 9279 | 1009 | 0.800 | 0.4725 | Yes | ||
| 18 | PFDN2 | 9551 | 1251 | 0.646 | 0.4723 | Yes | ||
| 19 | BAX | 17832 | 1302 | 0.620 | 0.4820 | Yes | ||
| 20 | RAMP2 | 20660 | 1417 | 0.562 | 0.4870 | Yes | ||
| 21 | CENTB5 | 15961 | 1563 | 0.506 | 0.4892 | Yes | ||
| 22 | ILF3 | 3110 3030 9176 | 1627 | 0.481 | 0.4954 | Yes | ||
| 23 | MRPL27 | 13573 | 1712 | 0.450 | 0.4998 | Yes | ||
| 24 | ATF7IP | 12028 13038 | 1720 | 0.448 | 0.5083 | Yes | ||
| 25 | PANK3 | 20924 | 1724 | 0.446 | 0.5171 | Yes | ||
| 26 | CLN3 | 17635 | 1753 | 0.437 | 0.5242 | Yes | ||
| 27 | ARL3 | 23652 | 1826 | 0.403 | 0.5284 | Yes | ||
| 28 | RGL2 | 23289 | 1879 | 0.383 | 0.5332 | Yes | ||
| 29 | HTF9C | 4886 1678 | 1908 | 0.374 | 0.5391 | Yes | ||
| 30 | CSRP2BP | 2696 10458 | 2011 | 0.340 | 0.5403 | Yes | ||
| 31 | STMN1 | 9261 | 2099 | 0.313 | 0.5419 | Yes | ||
| 32 | HMOX1 | 18565 | 2124 | 0.306 | 0.5466 | Yes | ||
| 33 | NCKIPSD | 19306 | 2178 | 0.289 | 0.5495 | Yes | ||
| 34 | SMYD4 | 20781 | 2262 | 0.263 | 0.5503 | Yes | ||
| 35 | RNF44 | 8364 | 2353 | 0.239 | 0.5502 | No | ||
| 36 | IGF2R | 23123 | 2511 | 0.194 | 0.5455 | No | ||
| 37 | HOOK2 | 18541 | 2597 | 0.173 | 0.5444 | No | ||
| 38 | NFX1 | 2475 | 2662 | 0.163 | 0.5441 | No | ||
| 39 | STC2 | 5526 | 2832 | 0.132 | 0.5376 | No | ||
| 40 | STX6 | 14091 12213 | 2835 | 0.131 | 0.5401 | No | ||
| 41 | PTMA | 9657 | 2964 | 0.111 | 0.5354 | No | ||
| 42 | MYST3 | 18651 | 3040 | 0.101 | 0.5334 | No | ||
| 43 | MPV17 | 9407 3505 3543 | 3066 | 0.097 | 0.5340 | No | ||
| 44 | TGIF2 | 6013 6012 | 3127 | 0.091 | 0.5325 | No | ||
| 45 | SNTB2 | 3887 18476 | 3246 | 0.078 | 0.5277 | No | ||
| 46 | LZTS2 | 10362 | 3324 | 0.073 | 0.5249 | No | ||
| 47 | TCF7 | 1467 20466 | 3425 | 0.064 | 0.5208 | No | ||
| 48 | HOXB7 | 666 9110 | 3467 | 0.061 | 0.5198 | No | ||
| 49 | DUSP7 | 10661 | 3487 | 0.060 | 0.5200 | No | ||
| 50 | KRTCAP2 | 15541 | 3519 | 0.058 | 0.5195 | No | ||
| 51 | WEE1 | 18127 | 3528 | 0.057 | 0.5202 | No | ||
| 52 | SYT3 | 18264 | 3563 | 0.056 | 0.5194 | No | ||
| 53 | TRIB1 | 22467 | 3914 | 0.041 | 0.5013 | No | ||
| 54 | VPS16 | 2927 14845 2825 | 4005 | 0.038 | 0.4972 | No | ||
| 55 | MAX | 21034 | 4042 | 0.036 | 0.4960 | No | ||
| 56 | KCNE4 | 12196 | 4328 | 0.029 | 0.4811 | No | ||
| 57 | ANKRD17 | 13496 3474 8170 | 4482 | 0.026 | 0.4733 | No | ||
| 58 | HPGD | 4867 | 4598 | 0.024 | 0.4676 | No | ||
| 59 | BHLHB3 | 16935 | 4686 | 0.023 | 0.4633 | No | ||
| 60 | HNRPH2 | 12085 7082 | 4693 | 0.023 | 0.4634 | No | ||
| 61 | GTF2H1 | 4069 18236 | 5435 | 0.015 | 0.4236 | No | ||
| 62 | KCTD15 | 17864 | 5688 | 0.013 | 0.4102 | No | ||
| 63 | NPM1 | 1196 | 5715 | 0.012 | 0.4090 | No | ||
| 64 | RANBP1 | 9692 5357 | 5795 | 0.012 | 0.4050 | No | ||
| 65 | RAB3IL1 | 3723 13229 | 5844 | 0.012 | 0.4026 | No | ||
| 66 | SEPT3 | 6275 10775 | 5975 | 0.011 | 0.3958 | No | ||
| 67 | PCDHA10 | 8792 | 6070 | 0.010 | 0.3909 | No | ||
| 68 | FGF14 | 21716 3005 3463 | 6188 | 0.010 | 0.3848 | No | ||
| 69 | BDNF | 14926 2797 | 6229 | 0.010 | 0.3828 | No | ||
| 70 | NDUFS1 | 13940 | 6418 | 0.009 | 0.3728 | No | ||
| 71 | FGF5 | 16785 | 6501 | 0.009 | 0.3685 | No | ||
| 72 | SC5DL | 6165 | 6567 | 0.008 | 0.3652 | No | ||
| 73 | TLL1 | 10188 | 6644 | 0.008 | 0.3612 | No | ||
| 74 | IPO7 | 6130 | 6752 | 0.008 | 0.3556 | No | ||
| 75 | NR3C2 | 18562 13798 | 6901 | 0.007 | 0.3477 | No | ||
| 76 | EN1 | 387 14155 | 6944 | 0.007 | 0.3456 | No | ||
| 77 | HIF1A | 4850 | 7024 | 0.007 | 0.3414 | No | ||
| 78 | ARTN | 15783 2430 2532 | 7058 | 0.007 | 0.3398 | No | ||
| 79 | HOXD10 | 14983 | 7260 | 0.006 | 0.3290 | No | ||
| 80 | PER1 | 20827 | 7564 | 0.005 | 0.3127 | No | ||
| 81 | LTBR | 16993 | 7578 | 0.005 | 0.3121 | No | ||
| 82 | SLC16A1 | 15468 | 7712 | 0.005 | 0.3050 | No | ||
| 83 | SCHIP1 | 15569 | 7817 | 0.004 | 0.2994 | No | ||
| 84 | HOXC12 | 22343 | 8309 | 0.003 | 0.2729 | No | ||
| 85 | ATP6V1H | 14301 | 8776 | 0.002 | 0.2477 | No | ||
| 86 | GLA | 4362 | 8788 | 0.002 | 0.2471 | No | ||
| 87 | NEUROD2 | 20262 | 8856 | 0.002 | 0.2435 | No | ||
| 88 | POGK | 7739 | 8859 | 0.002 | 0.2434 | No | ||
| 89 | UBE2B | 10245 | 8953 | 0.001 | 0.2384 | No | ||
| 90 | HRH3 | 14317 | 9008 | 0.001 | 0.2355 | No | ||
| 91 | CGN | 12896 12897 7701 | 9159 | 0.001 | 0.2274 | No | ||
| 92 | HNRPD | 8645 | 9217 | 0.001 | 0.2244 | No | ||
| 93 | RNF12 | 5384 | 9510 | -0.000 | 0.2085 | No | ||
| 94 | SSR1 | 4211 | 9622 | -0.000 | 0.2025 | No | ||
| 95 | UTP14C | 9704 | 9965 | -0.001 | 0.1840 | No | ||
| 96 | UBXD3 | 15701 | 10015 | -0.001 | 0.1814 | No | ||
| 97 | HOXA1 | 1005 17152 | 10103 | -0.002 | 0.1767 | No | ||
| 98 | NRAS | 5191 | 10314 | -0.002 | 0.1654 | No | ||
| 99 | APLN | 24164 | 10655 | -0.003 | 0.1470 | No | ||
| 100 | IRS4 | 9183 4926 | 10847 | -0.003 | 0.1368 | No | ||
| 101 | ELOVL3 | 23809 | 10942 | -0.004 | 0.1317 | No | ||
| 102 | ADAM10 | 4336 | 10985 | -0.004 | 0.1295 | No | ||
| 103 | ZNF318 | 12198 7179 | 11046 | -0.004 | 0.1264 | No | ||
| 104 | SYT13 | 14942 | 11048 | -0.004 | 0.1264 | No | ||
| 105 | SLC1A7 | 16147 | 11334 | -0.005 | 0.1111 | No | ||
| 106 | VLDLR | 5858 3763 912 3682 | 11374 | -0.005 | 0.1090 | No | ||
| 107 | EIF4E | 15403 1827 8890 | 11390 | -0.005 | 0.1083 | No | ||
| 108 | RHOQ | 23142 | 11429 | -0.005 | 0.1064 | No | ||
| 109 | DNMT3A | 2167 21330 | 11521 | -0.005 | 0.1015 | No | ||
| 110 | NEUROD1 | 14550 | 11595 | -0.006 | 0.0977 | No | ||
| 111 | HOXA3 | 9108 1075 | 11998 | -0.007 | 0.0761 | No | ||
| 112 | WDR20 | 21258 | 12705 | -0.010 | 0.0380 | No | ||
| 113 | KLF4 | 9230 4962 4963 | 12763 | -0.011 | 0.0352 | No | ||
| 114 | SLC26A2 | 23427 | 12978 | -0.012 | 0.0238 | No | ||
| 115 | ATP7B | 18918 4426 4427 8641 | 13140 | -0.013 | 0.0153 | No | ||
| 116 | HOXC5 | 22340 | 13242 | -0.014 | 0.0101 | No | ||
| 117 | HOXB6 | 20688 | 13332 | -0.014 | 0.0056 | No | ||
| 118 | LIN28 | 15723 | 13363 | -0.015 | 0.0043 | No | ||
| 119 | NR5A1 | 14597 | 13788 | -0.019 | -0.0183 | No | ||
| 120 | CDH1 | 18479 | 13858 | -0.020 | -0.0217 | No | ||
| 121 | TAC1 | 17526 19853 | 13947 | -0.021 | -0.0260 | No | ||
| 122 | SOCS5 | 1619 23141 | 14031 | -0.022 | -0.0301 | No | ||
| 123 | TRIM46 | 1779 15281 | 14396 | -0.028 | -0.0492 | No | ||
| 124 | HOXA9 | 17147 9109 1024 | 14436 | -0.029 | -0.0508 | No | ||
| 125 | HNRPDL | 11947 3551 6954 | 14665 | -0.035 | -0.0624 | No | ||
| 126 | GTF2A1 | 2064 8187 8188 13512 | 14773 | -0.038 | -0.0674 | No | ||
| 127 | NPTX1 | 20126 | 14838 | -0.041 | -0.0701 | No | ||
| 128 | SFXN2 | 3707 8255 3678 13588 | 15077 | -0.050 | -0.0820 | No | ||
| 129 | TAF6L | 5923 | 15296 | -0.062 | -0.0926 | No | ||
| 130 | ETV1 | 4688 8925 8924 | 15345 | -0.065 | -0.0939 | No | ||
| 131 | EGLN2 | 17920 | 15419 | -0.071 | -0.0964 | No | ||
| 132 | AGRP | 18768 | 15584 | -0.085 | -0.1036 | No | ||
| 133 | RPL19 | 9736 | 15770 | -0.103 | -0.1116 | No | ||
| 134 | ABR | 8468 20346 1476 1429 | 15940 | -0.129 | -0.1182 | No | ||
| 135 | SLC31A2 | 9827 | 16049 | -0.148 | -0.1211 | No | ||
| 136 | HMGN2 | 9095 | 16174 | -0.179 | -0.1242 | No | ||
| 137 | MCOLN1 | 13583 | 16370 | -0.222 | -0.1303 | No | ||
| 138 | LAMP1 | 18679 | 16513 | -0.261 | -0.1328 | No | ||
| 139 | RAPGEF3 | 22146 | 16578 | -0.277 | -0.1308 | No | ||
| 140 | CHD4 | 8418 17281 4225 | 16588 | -0.283 | -0.1256 | No | ||
| 141 | HOXA7 | 4862 | 17020 | -0.439 | -0.1402 | No | ||
| 142 | GNB2 | 9026 | 17072 | -0.460 | -0.1338 | No | ||
| 143 | GADD45G | 21637 | 17257 | -0.526 | -0.1333 | No | ||
| 144 | GNA13 | 20617 | 17337 | -0.563 | -0.1264 | No | ||
| 145 | ZFX | 5984 | 17401 | -0.596 | -0.1179 | No | ||
| 146 | WDR23 | 22009 | 17402 | -0.598 | -0.1060 | No | ||
| 147 | JUP | 4937 | 17462 | -0.635 | -0.0966 | No | ||
| 148 | SIRT1 | 19745 | 17477 | -0.643 | -0.0845 | No | ||
| 149 | COMMD3 | 73 | 17530 | -0.674 | -0.0739 | No | ||
| 150 | AXUD1 | 18967 | 17546 | -0.688 | -0.0610 | No | ||
| 151 | KBTBD2 | 17137 | 17749 | -0.823 | -0.0556 | No | ||
| 152 | WDR1 | 16544 3592 | 17801 | -0.867 | -0.0411 | No | ||
| 153 | WBP2 | 20146 | 18046 | -1.139 | -0.0316 | No | ||
| 154 | SGK | 9809 3419 | 18560 | -3.134 | 0.0030 | No |