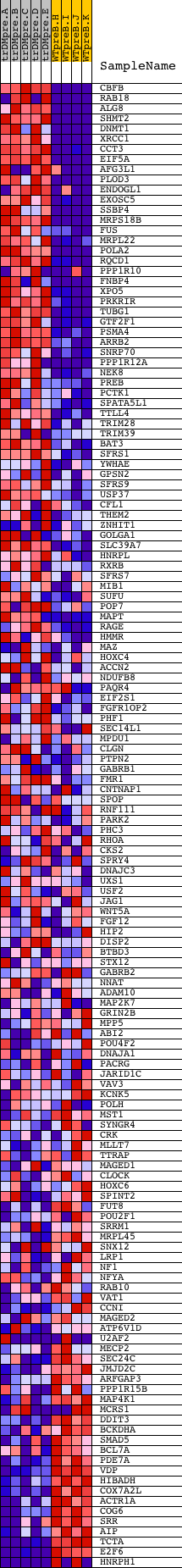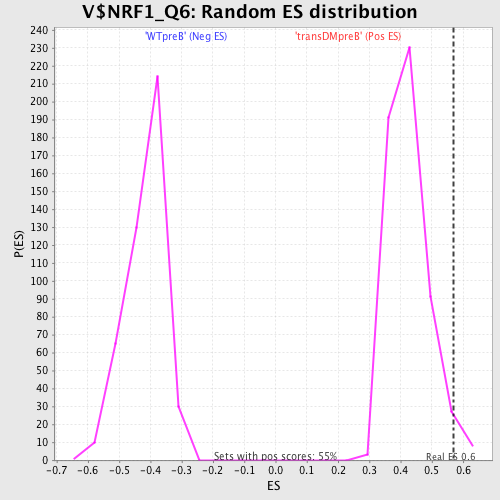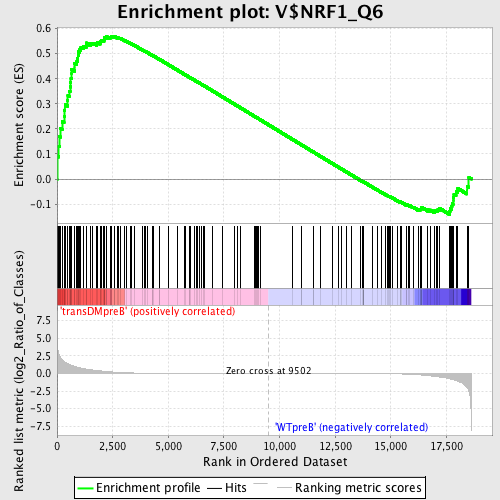
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | V$NRF1_Q6 |
| Enrichment Score (ES) | 0.5699194 |
| Normalized Enrichment Score (NES) | 1.3438725 |
| Nominal p-value | 0.027272727 |
| FDR q-value | 0.52408266 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CBFB | 3769 18499 | 3 | 5.871 | 0.0919 | Yes | ||
| 2 | RAB18 | 1972 5342 | 80 | 2.762 | 0.1311 | Yes | ||
| 3 | ALG8 | 11779 3912 2277 | 103 | 2.508 | 0.1692 | Yes | ||
| 4 | SHMT2 | 3307 19601 | 139 | 2.281 | 0.2031 | Yes | ||
| 5 | DNMT1 | 19217 | 226 | 1.914 | 0.2285 | Yes | ||
| 6 | XRCC1 | 173 | 312 | 1.633 | 0.2495 | Yes | ||
| 7 | CCT3 | 8709 | 321 | 1.612 | 0.2743 | Yes | ||
| 8 | EIF5A | 11345 20379 6590 | 354 | 1.574 | 0.2973 | Yes | ||
| 9 | AFG3L1 | 8547 4342 18427 3865 | 473 | 1.398 | 0.3128 | Yes | ||
| 10 | PLOD3 | 16666 | 486 | 1.369 | 0.3336 | Yes | ||
| 11 | ENDOGL1 | 3115 19275 | 543 | 1.285 | 0.3507 | Yes | ||
| 12 | EXOSC5 | 18334 | 585 | 1.210 | 0.3675 | Yes | ||
| 13 | SSBP4 | 13368 | 598 | 1.195 | 0.3856 | Yes | ||
| 14 | MRPS18B | 7365 | 622 | 1.171 | 0.4027 | Yes | ||
| 15 | FUS | 6135 | 643 | 1.147 | 0.4196 | Yes | ||
| 16 | MRPL22 | 10111 | 649 | 1.139 | 0.4372 | Yes | ||
| 17 | POLA2 | 23988 | 781 | 1.001 | 0.4458 | Yes | ||
| 18 | RQCD1 | 12205 | 782 | 0.999 | 0.4614 | Yes | ||
| 19 | PPP1R10 | 6972 11960 6971 | 885 | 0.905 | 0.4701 | Yes | ||
| 20 | FNBP4 | 14956 2671 | 924 | 0.876 | 0.4818 | Yes | ||
| 21 | XPO5 | 7802 | 938 | 0.860 | 0.4946 | Yes | ||
| 22 | PRKRIR | 7834 | 952 | 0.848 | 0.5072 | Yes | ||
| 23 | TUBG1 | 20662 | 1021 | 0.790 | 0.5159 | Yes | ||
| 24 | GTF2F1 | 13599 | 1068 | 0.756 | 0.5252 | Yes | ||
| 25 | PSMA4 | 11179 | 1176 | 0.687 | 0.5302 | Yes | ||
| 26 | ARRB2 | 20806 | 1303 | 0.619 | 0.5331 | Yes | ||
| 27 | SNRP70 | 1186 | 1306 | 0.617 | 0.5426 | Yes | ||
| 28 | PPP1R12A | 3354 19886 | 1508 | 0.525 | 0.5400 | Yes | ||
| 29 | NEK8 | 20338 | 1606 | 0.486 | 0.5424 | Yes | ||
| 30 | PREB | 6948 3628 | 1781 | 0.426 | 0.5396 | Yes | ||
| 31 | PCTK1 | 24369 | 1799 | 0.414 | 0.5452 | Yes | ||
| 32 | SPATA5L1 | 10062 | 1928 | 0.367 | 0.5440 | Yes | ||
| 33 | TTLL4 | 328 | 1962 | 0.356 | 0.5478 | Yes | ||
| 34 | TRIM28 | 18389 | 1977 | 0.352 | 0.5526 | Yes | ||
| 35 | TRIM39 | 22994 | 2080 | 0.319 | 0.5520 | Yes | ||
| 36 | BAT3 | 5896 | 2111 | 0.309 | 0.5553 | Yes | ||
| 37 | SFRS1 | 8492 | 2114 | 0.308 | 0.5600 | Yes | ||
| 38 | YWHAE | 20776 | 2133 | 0.303 | 0.5638 | Yes | ||
| 39 | GPSN2 | 18823 3868 | 2202 | 0.280 | 0.5645 | Yes | ||
| 40 | SFRS9 | 16731 | 2228 | 0.274 | 0.5674 | Yes | ||
| 41 | USP37 | 13923 | 2377 | 0.233 | 0.5630 | Yes | ||
| 42 | CFL1 | 4516 | 2394 | 0.228 | 0.5658 | Yes | ||
| 43 | THEM2 | 21512 | 2438 | 0.217 | 0.5668 | Yes | ||
| 44 | ZNHIT1 | 16334 | 2444 | 0.215 | 0.5699 | Yes | ||
| 45 | GOLGA1 | 14595 | 2560 | 0.183 | 0.5666 | No | ||
| 46 | SLC39A7 | 23022 | 2589 | 0.175 | 0.5678 | No | ||
| 47 | HNRPL | 18315 | 2700 | 0.157 | 0.5643 | No | ||
| 48 | RXRB | 23285 9768 | 2769 | 0.144 | 0.5629 | No | ||
| 49 | SFRS7 | 22889 | 2839 | 0.130 | 0.5612 | No | ||
| 50 | MIB1 | 5908 | 3038 | 0.101 | 0.5520 | No | ||
| 51 | SUFU | 6283 | 3137 | 0.090 | 0.5481 | No | ||
| 52 | POP7 | 16329 | 3298 | 0.074 | 0.5406 | No | ||
| 53 | MAPT | 9420 1347 | 3342 | 0.071 | 0.5394 | No | ||
| 54 | RAGE | 6465 2056 2165 | 3491 | 0.060 | 0.5323 | No | ||
| 55 | HMMR | 20493 1243 | 3835 | 0.044 | 0.5145 | No | ||
| 56 | MAZ | 1327 17623 | 3923 | 0.040 | 0.5104 | No | ||
| 57 | HOXC4 | 4864 | 3955 | 0.039 | 0.5093 | No | ||
| 58 | ACCN2 | 22363 | 4059 | 0.036 | 0.5043 | No | ||
| 59 | NDUFB8 | 7418 | 4266 | 0.031 | 0.4936 | No | ||
| 60 | PAQR4 | 23105 | 4294 | 0.030 | 0.4926 | No | ||
| 61 | EIF2S1 | 4658 | 4327 | 0.029 | 0.4914 | No | ||
| 62 | FGFR1OP2 | 12544 12543 7457 | 4615 | 0.024 | 0.4762 | No | ||
| 63 | PHF1 | 23324 1551 | 4618 | 0.024 | 0.4765 | No | ||
| 64 | SEC14L1 | 1212 20583 | 5003 | 0.019 | 0.4560 | No | ||
| 65 | MPDU1 | 20386 | 5397 | 0.015 | 0.4349 | No | ||
| 66 | CLGN | 4528 | 5718 | 0.012 | 0.4178 | No | ||
| 67 | PTPN2 | 23414 | 5771 | 0.012 | 0.4152 | No | ||
| 68 | GABRB1 | 16832 | 5932 | 0.011 | 0.4067 | No | ||
| 69 | FMR1 | 24321 | 5960 | 0.011 | 0.4054 | No | ||
| 70 | CNTNAP1 | 11987 | 6015 | 0.011 | 0.4026 | No | ||
| 71 | SPOP | 5498 | 6186 | 0.010 | 0.3936 | No | ||
| 72 | RNF111 | 13555 8218 13556 | 6272 | 0.009 | 0.3891 | No | ||
| 73 | PARK2 | 6944 11939 1499 23385 | 6311 | 0.009 | 0.3872 | No | ||
| 74 | PHC3 | 6313 | 6379 | 0.009 | 0.3837 | No | ||
| 75 | RHOA | 8624 4409 4410 | 6499 | 0.009 | 0.3774 | No | ||
| 76 | CKS2 | 7264 | 6565 | 0.008 | 0.3741 | No | ||
| 77 | SPRY4 | 23448 | 6618 | 0.008 | 0.3714 | No | ||
| 78 | DNAJC3 | 21934 | 7002 | 0.007 | 0.3507 | No | ||
| 79 | UXS1 | 13965 | 7434 | 0.005 | 0.3275 | No | ||
| 80 | USF2 | 10262 5833 10263 | 7969 | 0.004 | 0.2986 | No | ||
| 81 | JAG1 | 14415 | 8098 | 0.004 | 0.2918 | No | ||
| 82 | WNT5A | 22066 5880 | 8234 | 0.003 | 0.2845 | No | ||
| 83 | FGF12 | 1723 22621 | 8850 | 0.002 | 0.2512 | No | ||
| 84 | HIP2 | 3626 3485 16838 | 8926 | 0.001 | 0.2472 | No | ||
| 85 | DISP2 | 5652 | 8981 | 0.001 | 0.2443 | No | ||
| 86 | BTBD3 | 10457 2975 6007 6006 | 9028 | 0.001 | 0.2418 | No | ||
| 87 | STX12 | 4140 | 9058 | 0.001 | 0.2403 | No | ||
| 88 | GABRB2 | 20922 | 9150 | 0.001 | 0.2353 | No | ||
| 89 | NNAT | 2764 5180 | 10580 | -0.003 | 0.1580 | No | ||
| 90 | ADAM10 | 4336 | 10985 | -0.004 | 0.1362 | No | ||
| 91 | MAP2K7 | 6453 | 10987 | -0.004 | 0.1362 | No | ||
| 92 | GRIN2B | 16957 | 11537 | -0.005 | 0.1066 | No | ||
| 93 | MPP5 | 21230 7078 | 11541 | -0.005 | 0.1065 | No | ||
| 94 | ABI2 | 4037 6830 | 11849 | -0.006 | 0.0900 | No | ||
| 95 | POU4F2 | 9603 5276 | 12393 | -0.009 | 0.0607 | No | ||
| 96 | DNAJA1 | 4878 16242 2345 | 12649 | -0.010 | 0.0471 | No | ||
| 97 | PACRG | 23127 | 12800 | -0.011 | 0.0391 | No | ||
| 98 | JARID1C | 5460 2563 | 13024 | -0.012 | 0.0272 | No | ||
| 99 | VAV3 | 1774 1848 15443 | 13227 | -0.013 | 0.0165 | No | ||
| 100 | KCNK5 | 4946 9212 | 13632 | -0.017 | -0.0051 | No | ||
| 101 | POLH | 22966 | 13705 | -0.018 | -0.0087 | No | ||
| 102 | MST1 | 9089 | 13749 | -0.018 | -0.0108 | No | ||
| 103 | SYNGR4 | 17825 | 13761 | -0.018 | -0.0111 | No | ||
| 104 | CRK | 4559 1249 | 13772 | -0.019 | -0.0113 | No | ||
| 105 | MLLT7 | 24278 | 13782 | -0.019 | -0.0115 | No | ||
| 106 | TTRAP | 21692 | 14180 | -0.025 | -0.0326 | No | ||
| 107 | MAGED1 | 24108 | 14402 | -0.029 | -0.0441 | No | ||
| 108 | CLOCK | 8749 16507 4530 | 14582 | -0.033 | -0.0533 | No | ||
| 109 | HOXC6 | 22339 2302 | 14775 | -0.039 | -0.0631 | No | ||
| 110 | SPINT2 | 4128 17899 | 14834 | -0.040 | -0.0656 | No | ||
| 111 | FUT8 | 2163 21233 | 14891 | -0.043 | -0.0680 | No | ||
| 112 | POU2F1 | 5275 3989 4065 4010 | 14921 | -0.044 | -0.0688 | No | ||
| 113 | SRRM1 | 6958 | 14978 | -0.046 | -0.0712 | No | ||
| 114 | MRPL45 | 20680 | 15071 | -0.050 | -0.0753 | No | ||
| 115 | SNX12 | 7061 | 15311 | -0.062 | -0.0873 | No | ||
| 116 | LRP1 | 9284 | 15413 | -0.071 | -0.0917 | No | ||
| 117 | NF1 | 5165 | 15418 | -0.071 | -0.0908 | No | ||
| 118 | NFYA | 5172 | 15486 | -0.077 | -0.0932 | No | ||
| 119 | RAB10 | 2090 21119 | 15711 | -0.096 | -0.1038 | No | ||
| 120 | VAT1 | 11253 | 15722 | -0.097 | -0.1028 | No | ||
| 121 | CCNI | 4493 | 15806 | -0.107 | -0.1056 | No | ||
| 122 | MAGED2 | 24034 | 15859 | -0.114 | -0.1067 | No | ||
| 123 | ATP6V1D | 7872 | 16014 | -0.142 | -0.1128 | No | ||
| 124 | U2AF2 | 5813 5812 5814 | 16244 | -0.194 | -0.1221 | No | ||
| 125 | MECP2 | 5088 | 16314 | -0.209 | -0.1226 | No | ||
| 126 | SEC24C | 22086 | 16319 | -0.210 | -0.1195 | No | ||
| 127 | JMJD2C | 16188 | 16362 | -0.220 | -0.1183 | No | ||
| 128 | ARFGAP3 | 22183 | 16364 | -0.221 | -0.1149 | No | ||
| 129 | PPP1R15B | 4254 14135 | 16368 | -0.222 | -0.1116 | No | ||
| 130 | MAP4K1 | 18313 | 16669 | -0.306 | -0.1230 | No | ||
| 131 | MCRS1 | 6961 6960 6962 | 16776 | -0.342 | -0.1234 | No | ||
| 132 | DDIT3 | 19857 | 16966 | -0.420 | -0.1271 | No | ||
| 133 | BCKDHA | 17925 | 17032 | -0.442 | -0.1236 | No | ||
| 134 | SMAD5 | 21621 | 17117 | -0.477 | -0.1207 | No | ||
| 135 | BCL7A | 8063 | 17193 | -0.504 | -0.1169 | No | ||
| 136 | PDE7A | 9544 1788 | 17646 | -0.750 | -0.1296 | No | ||
| 137 | VDP | 16794 | 17662 | -0.764 | -0.1184 | No | ||
| 138 | HIBADH | 17144 | 17717 | -0.804 | -0.1087 | No | ||
| 139 | COX7A2L | 9819 | 17751 | -0.827 | -0.0976 | No | ||
| 140 | ACTR1A | 23653 | 17798 | -0.866 | -0.0865 | No | ||
| 141 | COG6 | 15345 | 17802 | -0.868 | -0.0730 | No | ||
| 142 | SRR | 6566 11328 | 17837 | -0.894 | -0.0608 | No | ||
| 143 | AIP | 23955 | 17931 | -0.997 | -0.0502 | No | ||
| 144 | TCTA | 4167 | 18012 | -1.092 | -0.0375 | No | ||
| 145 | E2F6 | 6925 6926 11920 | 18424 | -1.951 | -0.0291 | No | ||
| 146 | HNRPH1 | 12222 1486 1220 7202 7201 | 18508 | -2.516 | 0.0058 | No |