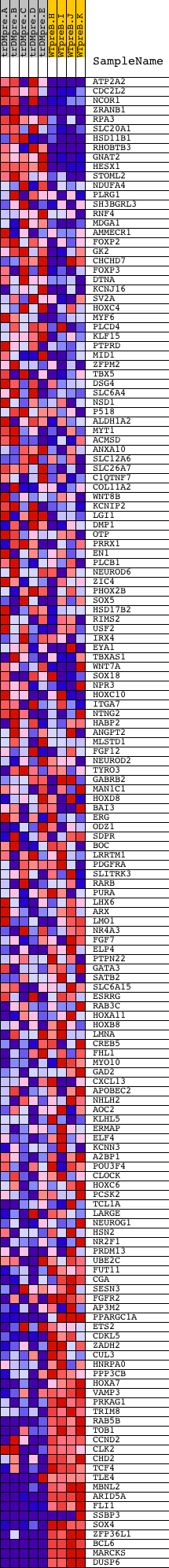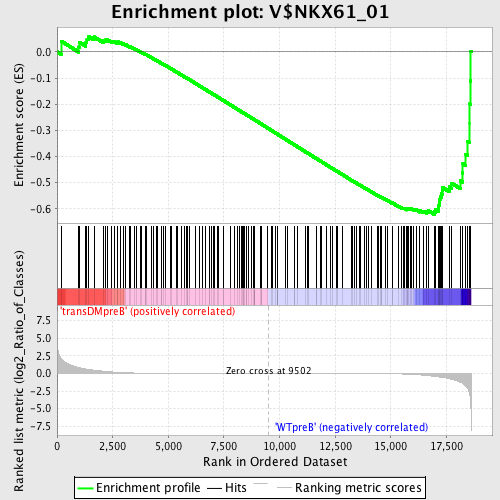
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$NKX61_01 |
| Enrichment Score (ES) | -0.62190336 |
| Normalized Enrichment Score (NES) | -1.5029912 |
| Nominal p-value | 0.0022222223 |
| FDR q-value | 0.14648347 |
| FWER p-Value | 0.783 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A2 | 4421 3481 | 184 | 2.086 | 0.0415 | No | ||
| 2 | CDC2L2 | 15964 2419 | 979 | 0.822 | 0.0187 | No | ||
| 3 | NCOR1 | 5402 | 1004 | 0.803 | 0.0372 | No | ||
| 4 | ZRANB1 | 6884 11702 | 1294 | 0.624 | 0.0369 | No | ||
| 5 | RPA3 | 12667 | 1341 | 0.601 | 0.0492 | No | ||
| 6 | SLC20A1 | 14855 | 1398 | 0.571 | 0.0603 | No | ||
| 7 | HSD11B1 | 9125 4019 | 1667 | 0.464 | 0.0572 | No | ||
| 8 | RHOBTB3 | 7849 | 2090 | 0.316 | 0.0421 | No | ||
| 9 | GNAT2 | 15454 | 2161 | 0.294 | 0.0456 | No | ||
| 10 | HESX1 | 22070 | 2270 | 0.261 | 0.0462 | No | ||
| 11 | STOML2 | 15902 | 2465 | 0.207 | 0.0407 | No | ||
| 12 | NDUFA4 | 9451 | 2580 | 0.178 | 0.0390 | No | ||
| 13 | PLRG1 | 15565 | 2716 | 0.154 | 0.0355 | No | ||
| 14 | SH3BGRL3 | 15722 | 2734 | 0.151 | 0.0383 | No | ||
| 15 | RNF4 | 5385 9729 | 2854 | 0.128 | 0.0350 | No | ||
| 16 | MDGA1 | 13231 | 2974 | 0.110 | 0.0312 | No | ||
| 17 | AMMECR1 | 7067 | 3094 | 0.095 | 0.0271 | No | ||
| 18 | FOXP2 | 4312 17523 8523 | 3267 | 0.076 | 0.0197 | No | ||
| 19 | GK2 | 16472 | 3315 | 0.073 | 0.0190 | No | ||
| 20 | CHCHD7 | 16280 | 3497 | 0.060 | 0.0106 | No | ||
| 21 | FOXP3 | 9804 5426 | 3549 | 0.056 | 0.0093 | No | ||
| 22 | DTNA | 8870 4647 23611 1991 | 3759 | 0.047 | -0.0009 | No | ||
| 23 | KCNJ16 | 20613 | 3783 | 0.046 | -0.0010 | No | ||
| 24 | SV2A | 15498 12250 | 3953 | 0.039 | -0.0092 | No | ||
| 25 | HOXC4 | 4864 | 3955 | 0.039 | -0.0083 | No | ||
| 26 | MYF6 | 19631 | 4018 | 0.037 | -0.0108 | No | ||
| 27 | PLCD4 | 9587 | 4245 | 0.031 | -0.0222 | No | ||
| 28 | KLF15 | 17359 | 4348 | 0.029 | -0.0270 | No | ||
| 29 | PTPRD | 11554 5330 | 4447 | 0.027 | -0.0317 | No | ||
| 30 | MID1 | 5097 5098 | 4528 | 0.025 | -0.0354 | No | ||
| 31 | ZFPM2 | 22483 | 4704 | 0.022 | -0.0443 | No | ||
| 32 | TBX5 | 3531 5634 | 4706 | 0.022 | -0.0438 | No | ||
| 33 | DSG4 | 9262 23616 | 4791 | 0.021 | -0.0479 | No | ||
| 34 | SLC6A4 | 20770 | 4859 | 0.021 | -0.0510 | No | ||
| 35 | NSD1 | 2134 5197 | 5089 | 0.018 | -0.0630 | No | ||
| 36 | P518 | 14624 | 5159 | 0.017 | -0.0663 | No | ||
| 37 | ALDH1A2 | 19386 | 5372 | 0.015 | -0.0774 | No | ||
| 38 | MYT1 | 36 | 5398 | 0.015 | -0.0784 | No | ||
| 39 | ACMSD | 14152 | 5589 | 0.013 | -0.0883 | No | ||
| 40 | ANXA10 | 18869 | 5705 | 0.012 | -0.0943 | No | ||
| 41 | SLC12A6 | 8413 2829 2874 | 5818 | 0.012 | -0.1000 | No | ||
| 42 | SLC26A7 | 5538 2372 | 5839 | 0.012 | -0.1008 | No | ||
| 43 | C1QTNF7 | 16852 | 5872 | 0.012 | -0.1023 | No | ||
| 44 | COL11A2 | 23286 | 5941 | 0.011 | -0.1057 | No | ||
| 45 | WNT8B | 23841 | 6216 | 0.010 | -0.1203 | No | ||
| 46 | KCNIP2 | 23655 3725 | 6388 | 0.009 | -0.1293 | No | ||
| 47 | LGI1 | 23867 | 6524 | 0.008 | -0.1364 | No | ||
| 48 | DMP1 | 16776 | 6682 | 0.008 | -0.1448 | No | ||
| 49 | OTP | 21582 | 6828 | 0.007 | -0.1524 | No | ||
| 50 | PRRX1 | 9595 5271 | 6850 | 0.007 | -0.1534 | No | ||
| 51 | EN1 | 387 14155 | 6944 | 0.007 | -0.1583 | No | ||
| 52 | PLCB1 | 14832 2821 | 7050 | 0.007 | -0.1638 | No | ||
| 53 | NEUROD6 | 17138 | 7095 | 0.006 | -0.1660 | No | ||
| 54 | ZIC4 | 5992 | 7199 | 0.006 | -0.1714 | No | ||
| 55 | PHOX2B | 16524 | 7272 | 0.006 | -0.1752 | No | ||
| 56 | SOX5 | 9850 1044 5480 16937 | 7475 | 0.005 | -0.1860 | No | ||
| 57 | HSD17B2 | 18452 | 7490 | 0.005 | -0.1866 | No | ||
| 58 | RIMS2 | 4370 | 7789 | 0.004 | -0.2027 | No | ||
| 59 | USF2 | 10262 5833 10263 | 7969 | 0.004 | -0.2123 | No | ||
| 60 | IRX4 | 11944 | 8121 | 0.003 | -0.2204 | No | ||
| 61 | EYA1 | 4695 4061 | 8207 | 0.003 | -0.2249 | No | ||
| 62 | TBXAS1 | 17480 1105 | 8273 | 0.003 | -0.2283 | No | ||
| 63 | WNT7A | 17068 | 8335 | 0.003 | -0.2316 | No | ||
| 64 | SOX18 | 14302 | 8371 | 0.003 | -0.2334 | No | ||
| 65 | NPR3 | 5188 | 8396 | 0.003 | -0.2346 | No | ||
| 66 | HOXC10 | 22342 | 8431 | 0.003 | -0.2364 | No | ||
| 67 | ITGA7 | 19835 | 8493 | 0.002 | -0.2397 | No | ||
| 68 | NTNG2 | 2731 9330 | 8514 | 0.002 | -0.2407 | No | ||
| 69 | HABP2 | 10364 23825 | 8591 | 0.002 | -0.2447 | No | ||
| 70 | ANGPT2 | 14885 3783 18923 | 8731 | 0.002 | -0.2522 | No | ||
| 71 | MLSTD1 | 17234 | 8805 | 0.002 | -0.2561 | No | ||
| 72 | FGF12 | 1723 22621 | 8850 | 0.002 | -0.2585 | No | ||
| 73 | NEUROD2 | 20262 | 8856 | 0.002 | -0.2587 | No | ||
| 74 | TYRO3 | 5811 | 8858 | 0.002 | -0.2587 | No | ||
| 75 | GABRB2 | 20922 | 9150 | 0.001 | -0.2745 | No | ||
| 76 | MAN1C1 | 10514 6064 15714 | 9188 | 0.001 | -0.2765 | No | ||
| 77 | HOXD8 | 9116 | 9193 | 0.001 | -0.2767 | No | ||
| 78 | BAI3 | 5580 | 9437 | 0.000 | -0.2898 | No | ||
| 79 | ERG | 1686 8915 | 9637 | -0.000 | -0.3006 | No | ||
| 80 | ODZ1 | 24168 | 9659 | -0.000 | -0.3017 | No | ||
| 81 | SDPR | 14252 | 9796 | -0.001 | -0.3091 | No | ||
| 82 | BOC | 22587 | 9893 | -0.001 | -0.3142 | No | ||
| 83 | LRRTM1 | 17408 | 9895 | -0.001 | -0.3143 | No | ||
| 84 | PDGFRA | 16824 | 9909 | -0.001 | -0.3149 | No | ||
| 85 | SLITRK3 | 11816 | 10256 | -0.002 | -0.3336 | No | ||
| 86 | RARB | 3011 21915 | 10377 | -0.002 | -0.3401 | No | ||
| 87 | PURA | 9670 | 10677 | -0.003 | -0.3562 | No | ||
| 88 | LHX6 | 9275 2914 | 10689 | -0.003 | -0.3567 | No | ||
| 89 | ARX | 24290 | 10825 | -0.003 | -0.3640 | No | ||
| 90 | LMO1 | 17685 | 11162 | -0.004 | -0.3821 | No | ||
| 91 | NR4A3 | 9473 16212 5183 | 11248 | -0.004 | -0.3866 | No | ||
| 92 | FGF7 | 8965 | 11293 | -0.005 | -0.3888 | No | ||
| 93 | ELP4 | 8094 | 11639 | -0.006 | -0.4074 | No | ||
| 94 | PTPN22 | 1775 15470 | 11642 | -0.006 | -0.4073 | No | ||
| 95 | GATA3 | 9004 | 11845 | -0.006 | -0.4181 | No | ||
| 96 | SATB2 | 5604 | 11896 | -0.007 | -0.4207 | No | ||
| 97 | SLC6A15 | 8335 3321 | 12125 | -0.007 | -0.4328 | No | ||
| 98 | ESRRG | 6447 | 12271 | -0.008 | -0.4405 | No | ||
| 99 | RAB3C | 21351 | 12385 | -0.009 | -0.4464 | No | ||
| 100 | HOXA11 | 17146 | 12539 | -0.009 | -0.4545 | No | ||
| 101 | HOXB8 | 9111 | 12624 | -0.010 | -0.4588 | No | ||
| 102 | LMNA | 4999 15290 1778 9280 | 12841 | -0.011 | -0.4702 | No | ||
| 103 | CREB5 | 10551 | 13229 | -0.013 | -0.4908 | No | ||
| 104 | FHL1 | 24334 | 13271 | -0.014 | -0.4927 | No | ||
| 105 | MYO10 | 2184 5141 | 13360 | -0.014 | -0.4971 | No | ||
| 106 | GAD2 | 15108 | 13436 | -0.015 | -0.5008 | No | ||
| 107 | CXCL13 | 16789 | 13588 | -0.017 | -0.5086 | No | ||
| 108 | APOBEC2 | 22946 | 13633 | -0.017 | -0.5106 | No | ||
| 109 | NHLH2 | 9462 | 13802 | -0.019 | -0.5192 | No | ||
| 110 | AOC2 | 20658 1256 | 13888 | -0.020 | -0.5233 | No | ||
| 111 | KLHL5 | 3631 7753 | 14003 | -0.022 | -0.5289 | No | ||
| 112 | ERMAP | 15776 | 14121 | -0.023 | -0.5347 | No | ||
| 113 | ELF4 | 24162 | 14390 | -0.028 | -0.5485 | No | ||
| 114 | KCNN3 | 15538 1913 | 14433 | -0.029 | -0.5501 | No | ||
| 115 | A2BP1 | 11212 1652 1689 | 14530 | -0.031 | -0.5545 | No | ||
| 116 | POU3F4 | 347 | 14572 | -0.033 | -0.5559 | No | ||
| 117 | CLOCK | 8749 16507 4530 | 14582 | -0.033 | -0.5556 | No | ||
| 118 | HOXC6 | 22339 2302 | 14775 | -0.039 | -0.5650 | No | ||
| 119 | PCSK2 | 9537 5231 14824 | 14833 | -0.040 | -0.5671 | No | ||
| 120 | TCL1A | 20987 | 15062 | -0.050 | -0.5783 | No | ||
| 121 | LARGE | 18837 | 15067 | -0.050 | -0.5773 | No | ||
| 122 | NEUROG1 | 21446 | 15346 | -0.065 | -0.5907 | No | ||
| 123 | HSN2 | 441 | 15474 | -0.076 | -0.5957 | No | ||
| 124 | NR2F1 | 7015 21401 | 15561 | -0.083 | -0.5984 | No | ||
| 125 | PRDM13 | 15933 | 15593 | -0.085 | -0.5979 | No | ||
| 126 | UBE2C | 12714 | 15720 | -0.097 | -0.6024 | No | ||
| 127 | FUT11 | 13084 | 15733 | -0.099 | -0.6006 | No | ||
| 128 | CGA | 16246 | 15768 | -0.103 | -0.5999 | No | ||
| 129 | SESN3 | 19561 | 15793 | -0.106 | -0.5986 | No | ||
| 130 | FGFR2 | 1917 4722 1119 | 15861 | -0.114 | -0.5994 | No | ||
| 131 | AP3M2 | 11 | 15927 | -0.126 | -0.5998 | No | ||
| 132 | PPARGC1A | 16533 | 16024 | -0.143 | -0.6015 | No | ||
| 133 | ETS2 | 10716 | 16147 | -0.172 | -0.6038 | No | ||
| 134 | CDKL5 | 24019 | 16309 | -0.208 | -0.6074 | No | ||
| 135 | ZADH2 | 23501 | 16450 | -0.240 | -0.6091 | No | ||
| 136 | CUL3 | 6470 | 16616 | -0.291 | -0.6109 | No | ||
| 137 | HNRPA0 | 13382 | 16677 | -0.308 | -0.6065 | No | ||
| 138 | PPP3CB | 5285 | 16962 | -0.419 | -0.6116 | Yes | ||
| 139 | HOXA7 | 4862 | 17020 | -0.439 | -0.6039 | Yes | ||
| 140 | VAMP3 | 15660 | 17144 | -0.486 | -0.5986 | Yes | ||
| 141 | PRKAG1 | 22135 | 17156 | -0.489 | -0.5871 | Yes | ||
| 142 | TRIM8 | 23824 | 17173 | -0.497 | -0.5757 | Yes | ||
| 143 | RAB5B | 359 19591 3430 | 17180 | -0.501 | -0.5637 | Yes | ||
| 144 | TOB1 | 20703 | 17217 | -0.513 | -0.5530 | Yes | ||
| 145 | CCND2 | 16987 | 17264 | -0.529 | -0.5424 | Yes | ||
| 146 | CLK2 | 1883 15544 | 17311 | -0.551 | -0.5313 | Yes | ||
| 147 | CHD2 | 10847 | 17320 | -0.556 | -0.5180 | Yes | ||
| 148 | TCF4 | 5642 | 17625 | -0.736 | -0.5164 | Yes | ||
| 149 | TLE4 | 23720 3697 | 17739 | -0.818 | -0.5023 | Yes | ||
| 150 | MBNL2 | 21932 | 18117 | -1.242 | -0.4921 | Yes | ||
| 151 | ARID5A | 5668 | 18229 | -1.425 | -0.4630 | Yes | ||
| 152 | FLI1 | 4729 | 18239 | -1.444 | -0.4279 | Yes | ||
| 153 | SSBP3 | 2439 7814 2431 | 18373 | -1.803 | -0.3907 | Yes | ||
| 154 | SOX4 | 5479 | 18455 | -2.118 | -0.3428 | Yes | ||
| 155 | ZFP36L1 | 4453 4454 | 18552 | -2.980 | -0.2746 | Yes | ||
| 156 | BCL6 | 22624 | 18554 | -3.060 | -0.1992 | Yes | ||
| 157 | MARCKS | 9331 | 18574 | -3.642 | -0.1105 | Yes | ||
| 158 | DUSP6 | 19891 3399 | 18597 | -4.571 | 0.0010 | Yes |