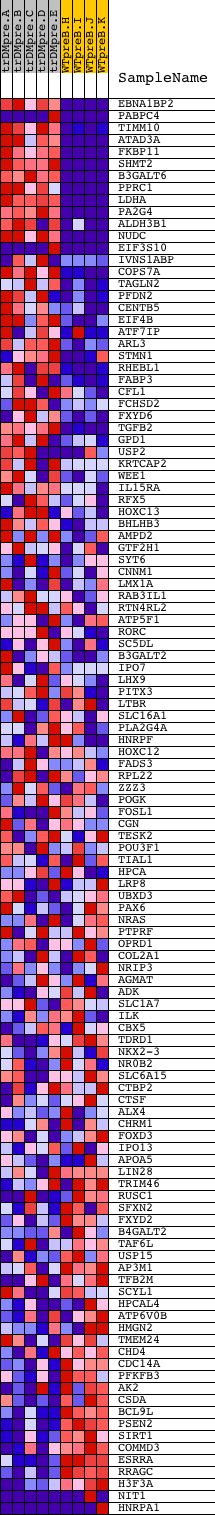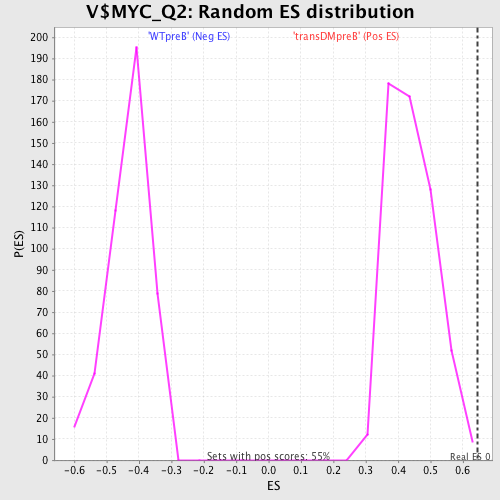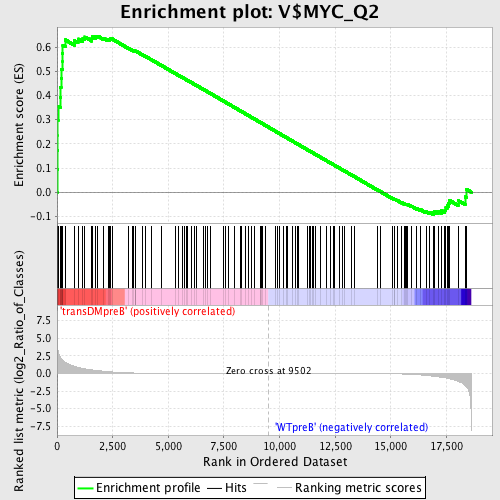
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.64576006 |
| Normalized Enrichment Score (NES) | 1.4658482 |
| Nominal p-value | 0.007259528 |
| FDR q-value | 0.48473337 |
| FWER p-Value | 0.952 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EBNA1BP2 | 8112 7599 12770 | 6 | 5.130 | 0.0949 | Yes | ||
| 2 | PABPC4 | 10510 2418 2548 | 17 | 4.160 | 0.1716 | Yes | ||
| 3 | TIMM10 | 11390 2158 | 30 | 3.408 | 0.2342 | Yes | ||
| 4 | ATAD3A | 8440 | 34 | 3.368 | 0.2966 | Yes | ||
| 5 | FKBP11 | 22136 | 44 | 3.221 | 0.3559 | Yes | ||
| 6 | SHMT2 | 3307 19601 | 139 | 2.281 | 0.3931 | Yes | ||
| 7 | B3GALT6 | 4391 | 153 | 2.224 | 0.4337 | Yes | ||
| 8 | PPRC1 | 23808 | 179 | 2.097 | 0.4713 | Yes | ||
| 9 | LDHA | 9269 | 190 | 2.057 | 0.5089 | Yes | ||
| 10 | PA2G4 | 5265 | 241 | 1.855 | 0.5407 | Yes | ||
| 11 | ALDH3B1 | 12569 23949 | 247 | 1.838 | 0.5745 | Yes | ||
| 12 | NUDC | 9495 | 260 | 1.798 | 0.6072 | Yes | ||
| 13 | EIF3S10 | 4659 8887 | 368 | 1.548 | 0.6302 | Yes | ||
| 14 | IVNS1ABP | 4384 4050 | 780 | 1.002 | 0.6266 | Yes | ||
| 15 | COPS7A | 11221 16997 | 941 | 0.857 | 0.6338 | Yes | ||
| 16 | TAGLN2 | 14043 | 1159 | 0.698 | 0.6351 | Yes | ||
| 17 | PFDN2 | 9551 | 1251 | 0.646 | 0.6421 | Yes | ||
| 18 | CENTB5 | 15961 | 1563 | 0.506 | 0.6347 | Yes | ||
| 19 | EIF4B | 13279 7979 | 1574 | 0.501 | 0.6435 | Yes | ||
| 20 | ATF7IP | 12028 13038 | 1720 | 0.448 | 0.6440 | Yes | ||
| 21 | ARL3 | 23652 | 1826 | 0.403 | 0.6458 | Yes | ||
| 22 | STMN1 | 9261 | 2099 | 0.313 | 0.6369 | No | ||
| 23 | RHEBL1 | 12781 | 2298 | 0.254 | 0.6309 | No | ||
| 24 | FABP3 | 8945 | 2372 | 0.233 | 0.6313 | No | ||
| 25 | CFL1 | 4516 | 2394 | 0.228 | 0.6344 | No | ||
| 26 | FCHSD2 | 18172 | 2468 | 0.205 | 0.6342 | No | ||
| 27 | FXYD6 | 19471 | 3215 | 0.081 | 0.5954 | No | ||
| 28 | TGFB2 | 5743 | 3382 | 0.068 | 0.5877 | No | ||
| 29 | GPD1 | 22361 | 3454 | 0.062 | 0.5850 | No | ||
| 30 | USP2 | 19480 3052 3043 | 3517 | 0.059 | 0.5827 | No | ||
| 31 | KRTCAP2 | 15541 | 3519 | 0.058 | 0.5838 | No | ||
| 32 | WEE1 | 18127 | 3528 | 0.057 | 0.5844 | No | ||
| 33 | IL15RA | 2899 2717 15118 2792 | 3833 | 0.044 | 0.5688 | No | ||
| 34 | RFX5 | 7018 | 3989 | 0.038 | 0.5611 | No | ||
| 35 | HOXC13 | 22344 | 4229 | 0.031 | 0.5488 | No | ||
| 36 | BHLHB3 | 16935 | 4686 | 0.023 | 0.5245 | No | ||
| 37 | AMPD2 | 1931 15197 | 5330 | 0.015 | 0.4901 | No | ||
| 38 | GTF2H1 | 4069 18236 | 5435 | 0.015 | 0.4847 | No | ||
| 39 | SYT6 | 13437 7042 12040 | 5623 | 0.013 | 0.4749 | No | ||
| 40 | CNNM1 | 23846 | 5720 | 0.012 | 0.4699 | No | ||
| 41 | LMX1A | 14062 3939 | 5829 | 0.012 | 0.4643 | No | ||
| 42 | RAB3IL1 | 3723 13229 | 5844 | 0.012 | 0.4637 | No | ||
| 43 | RTN4RL2 | 14544 | 6032 | 0.011 | 0.4538 | No | ||
| 44 | ATP5F1 | 15212 | 6185 | 0.010 | 0.4458 | No | ||
| 45 | RORC | 9732 | 6255 | 0.010 | 0.4422 | No | ||
| 46 | SC5DL | 6165 | 6567 | 0.008 | 0.4256 | No | ||
| 47 | B3GALT2 | 6498 | 6661 | 0.008 | 0.4207 | No | ||
| 48 | IPO7 | 6130 | 6752 | 0.008 | 0.4160 | No | ||
| 49 | LHX9 | 9276 | 6875 | 0.007 | 0.4095 | No | ||
| 50 | PITX3 | 9571 | 7464 | 0.005 | 0.3778 | No | ||
| 51 | LTBR | 16993 | 7578 | 0.005 | 0.3718 | No | ||
| 52 | SLC16A1 | 15468 | 7712 | 0.005 | 0.3647 | No | ||
| 53 | PLA2G4A | 13809 | 7986 | 0.004 | 0.3500 | No | ||
| 54 | HNRPF | 13607 | 8228 | 0.003 | 0.3370 | No | ||
| 55 | HOXC12 | 22343 | 8309 | 0.003 | 0.3328 | No | ||
| 56 | FADS3 | 7214 | 8485 | 0.003 | 0.3234 | No | ||
| 57 | RPL22 | 15979 | 8609 | 0.002 | 0.3168 | No | ||
| 58 | ZZZ3 | 15389 8445 4253 8444 1761 | 8722 | 0.002 | 0.3107 | No | ||
| 59 | POGK | 7739 | 8859 | 0.002 | 0.3034 | No | ||
| 60 | FOSL1 | 23779 | 9119 | 0.001 | 0.2894 | No | ||
| 61 | CGN | 12896 12897 7701 | 9159 | 0.001 | 0.2873 | No | ||
| 62 | TESK2 | 10504 | 9170 | 0.001 | 0.2868 | No | ||
| 63 | POU3F1 | 16093 | 9252 | 0.001 | 0.2824 | No | ||
| 64 | TIAL1 | 996 5753 | 9368 | 0.000 | 0.2762 | No | ||
| 65 | HPCA | 9118 | 9833 | -0.001 | 0.2512 | No | ||
| 66 | LRP8 | 16149 2425 | 9925 | -0.001 | 0.2463 | No | ||
| 67 | UBXD3 | 15701 | 10015 | -0.001 | 0.2415 | No | ||
| 68 | PAX6 | 5223 | 10181 | -0.002 | 0.2326 | No | ||
| 69 | NRAS | 5191 | 10314 | -0.002 | 0.2255 | No | ||
| 70 | PTPRF | 5331 2545 | 10372 | -0.002 | 0.2225 | No | ||
| 71 | OPRD1 | 15737 | 10577 | -0.003 | 0.2115 | No | ||
| 72 | COL2A1 | 22145 4542 | 10712 | -0.003 | 0.2043 | No | ||
| 73 | NRIP3 | 17679 | 10791 | -0.003 | 0.2001 | No | ||
| 74 | AGMAT | 16002 | 10844 | -0.003 | 0.1974 | No | ||
| 75 | ADK | 3302 3454 8555 | 11260 | -0.005 | 0.1750 | No | ||
| 76 | SLC1A7 | 16147 | 11334 | -0.005 | 0.1712 | No | ||
| 77 | ILK | 9177 1618 | 11394 | -0.005 | 0.1681 | No | ||
| 78 | CBX5 | 22108 4485 | 11480 | -0.005 | 0.1636 | No | ||
| 79 | TDRD1 | 13510 23818 | 11533 | -0.005 | 0.1609 | No | ||
| 80 | NKX2-3 | 5177 23845 | 11619 | -0.006 | 0.1564 | No | ||
| 81 | NR0B2 | 16050 | 11853 | -0.006 | 0.1439 | No | ||
| 82 | SLC6A15 | 8335 3321 | 12125 | -0.007 | 0.1294 | No | ||
| 83 | CTBP2 | 17591 1137 | 12274 | -0.008 | 0.1216 | No | ||
| 84 | CTSF | 23772 | 12416 | -0.009 | 0.1141 | No | ||
| 85 | ALX4 | 4379 | 12454 | -0.009 | 0.1123 | No | ||
| 86 | CHRM1 | 23943 | 12673 | -0.010 | 0.1007 | No | ||
| 87 | FOXD3 | 9088 | 12809 | -0.011 | 0.0936 | No | ||
| 88 | IPO13 | 15784 | 12914 | -0.011 | 0.0882 | No | ||
| 89 | APOA5 | 19469 | 13213 | -0.013 | 0.0723 | No | ||
| 90 | LIN28 | 15723 | 13363 | -0.015 | 0.0645 | No | ||
| 91 | TRIM46 | 1779 15281 | 14396 | -0.028 | 0.0092 | No | ||
| 92 | RUSC1 | 1853 15283 | 14532 | -0.031 | 0.0025 | No | ||
| 93 | SFXN2 | 3707 8255 3678 13588 | 15077 | -0.050 | -0.0259 | No | ||
| 94 | FXYD2 | 3038 19470 3068 3039 | 15144 | -0.053 | -0.0285 | No | ||
| 95 | B4GALT2 | 11990 | 15171 | -0.055 | -0.0289 | No | ||
| 96 | TAF6L | 5923 | 15296 | -0.062 | -0.0345 | No | ||
| 97 | USP15 | 4758 3377 | 15490 | -0.078 | -0.0435 | No | ||
| 98 | AP3M1 | 7059 | 15611 | -0.087 | -0.0483 | No | ||
| 99 | TFB2M | 4024 9092 4083 | 15652 | -0.090 | -0.0488 | No | ||
| 100 | SCYL1 | 23986 | 15704 | -0.096 | -0.0498 | No | ||
| 101 | HPCAL4 | 16096 | 15763 | -0.102 | -0.0510 | No | ||
| 102 | ATP6V0B | 15786 | 15928 | -0.126 | -0.0576 | No | ||
| 103 | HMGN2 | 9095 | 16174 | -0.179 | -0.0675 | No | ||
| 104 | TMEM24 | 19150 | 16342 | -0.214 | -0.0725 | No | ||
| 105 | CHD4 | 8418 17281 4225 | 16588 | -0.283 | -0.0805 | No | ||
| 106 | CDC14A | 15184 | 16732 | -0.329 | -0.0822 | No | ||
| 107 | PFKFB3 | 2748 | 16918 | -0.398 | -0.0848 | No | ||
| 108 | AK2 | 8563 2479 16073 | 16960 | -0.419 | -0.0792 | No | ||
| 109 | CSDA | 1163 16966 1134 | 17122 | -0.477 | -0.0791 | No | ||
| 110 | BCL9L | 19475 | 17265 | -0.530 | -0.0769 | No | ||
| 111 | PSEN2 | 13736 | 17430 | -0.615 | -0.0743 | No | ||
| 112 | SIRT1 | 19745 | 17477 | -0.643 | -0.0649 | No | ||
| 113 | COMMD3 | 73 | 17530 | -0.674 | -0.0552 | No | ||
| 114 | ESRRA | 23802 | 17590 | -0.710 | -0.0452 | No | ||
| 115 | RRAGC | 12015 | 17634 | -0.742 | -0.0338 | No | ||
| 116 | H3F3A | 4838 | 18026 | -1.107 | -0.0343 | No | ||
| 117 | NIT1 | 11293 933 | 18342 | -1.725 | -0.0193 | No | ||
| 118 | HNRPA1 | 9103 4860 | 18392 | -1.837 | 0.0121 | No |