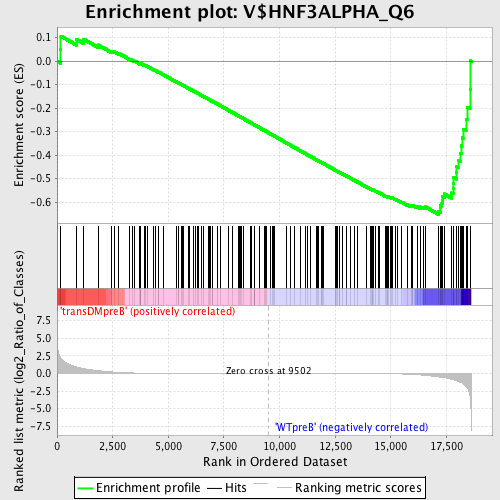
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$HNF3ALPHA_Q6 |
| Enrichment Score (ES) | -0.6523227 |
| Normalized Enrichment Score (NES) | -1.558394 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1340087 |
| FWER p-Value | 0.486 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FGF13 | 4720 8963 2627 | 160 | 2.208 | 0.0503 | No | ||
| 2 | ACACA | 309 4209 | 172 | 2.135 | 0.1067 | No | ||
| 3 | TAF5L | 18712 | 883 | 0.907 | 0.0925 | No | ||
| 4 | XPO4 | 7161 | 1195 | 0.677 | 0.0937 | No | ||
| 5 | TPP2 | 14257 | 1848 | 0.395 | 0.0690 | No | ||
| 6 | STOML2 | 15902 | 2465 | 0.207 | 0.0411 | No | ||
| 7 | GOLGA1 | 14595 | 2560 | 0.183 | 0.0409 | No | ||
| 8 | MEF2C | 3204 9378 | 2762 | 0.145 | 0.0339 | No | ||
| 9 | FOXP2 | 4312 17523 8523 | 3267 | 0.076 | 0.0087 | No | ||
| 10 | TWIST1 | 21291 | 3386 | 0.067 | 0.0041 | No | ||
| 11 | CHCHD7 | 16280 | 3497 | 0.060 | -0.0003 | No | ||
| 12 | SLC39A8 | 12547 1776 | 3723 | 0.048 | -0.0112 | No | ||
| 13 | H2AFY | 21447 | 3733 | 0.048 | -0.0104 | No | ||
| 14 | PALMD | 15181 | 3741 | 0.048 | -0.0095 | No | ||
| 15 | BCL11B | 7190 | 3768 | 0.046 | -0.0097 | No | ||
| 16 | TRIB1 | 22467 | 3914 | 0.041 | -0.0164 | No | ||
| 17 | HOXC4 | 4864 | 3955 | 0.039 | -0.0175 | No | ||
| 18 | TBX4 | 5633 | 4080 | 0.035 | -0.0233 | No | ||
| 19 | ATOH8 | 17120 | 4346 | 0.029 | -0.0369 | No | ||
| 20 | CP | 4555 | 4419 | 0.027 | -0.0401 | No | ||
| 21 | HOXB3 | 20685 | 4565 | 0.025 | -0.0472 | No | ||
| 22 | COL13A1 | 8763 | 4569 | 0.025 | -0.0468 | No | ||
| 23 | CYR61 | 9159 15145 9311 5024 | 4785 | 0.022 | -0.0578 | No | ||
| 24 | ELAVL4 | 15805 4889 9137 | 5378 | 0.015 | -0.0895 | No | ||
| 25 | GATA6 | 96 | 5459 | 0.014 | -0.0934 | No | ||
| 26 | NKD1 | 8228 | 5474 | 0.014 | -0.0938 | No | ||
| 27 | REPS2 | 24018 | 5580 | 0.013 | -0.0991 | No | ||
| 28 | NEBL | 14678 2691 | 5648 | 0.013 | -0.1024 | No | ||
| 29 | KCTD15 | 17864 | 5688 | 0.013 | -0.1042 | No | ||
| 30 | GCNT2 | 9168 3166 3243 21661 3270 | 5907 | 0.011 | -0.1157 | No | ||
| 31 | POGZ | 6031 1911 | 5936 | 0.011 | -0.1169 | No | ||
| 32 | SYN3 | 19658 | 6133 | 0.010 | -0.1272 | No | ||
| 33 | IRX5 | 18527 | 6203 | 0.010 | -0.1307 | No | ||
| 34 | SMPX | 24220 | 6329 | 0.009 | -0.1372 | No | ||
| 35 | CRH | 8784 | 6342 | 0.009 | -0.1376 | No | ||
| 36 | RBP3 | 22047 | 6489 | 0.009 | -0.1453 | No | ||
| 37 | ATOH1 | 4419 | 6594 | 0.008 | -0.1507 | No | ||
| 38 | PIK3C2A | 17667 | 6787 | 0.007 | -0.1609 | No | ||
| 39 | OTP | 21582 | 6828 | 0.007 | -0.1628 | No | ||
| 40 | PPP1CB | 5282 3529 3504 | 6891 | 0.007 | -0.1660 | No | ||
| 41 | PTPRG | 22097 3151 | 6990 | 0.007 | -0.1711 | No | ||
| 42 | HSF2 | 9128 | 7001 | 0.007 | -0.1715 | No | ||
| 43 | PITX2 | 15424 1878 | 7190 | 0.006 | -0.1815 | No | ||
| 44 | SOX15 | 9848 | 7334 | 0.006 | -0.1891 | No | ||
| 45 | TNR | 5787 | 7722 | 0.004 | -0.2099 | No | ||
| 46 | WNT4 | 16025 | 7869 | 0.004 | -0.2177 | No | ||
| 47 | GDF8 | 9411 | 8130 | 0.003 | -0.2317 | No | ||
| 48 | BOK | 11953 14174 | 8180 | 0.003 | -0.2343 | No | ||
| 49 | WNT5A | 22066 5880 | 8234 | 0.003 | -0.2370 | No | ||
| 50 | TBXAS1 | 17480 1105 | 8273 | 0.003 | -0.2390 | No | ||
| 51 | MYL1 | 4015 | 8370 | 0.003 | -0.2441 | No | ||
| 52 | KLF12 | 4960 2637 21732 9228 | 8710 | 0.002 | -0.2624 | No | ||
| 53 | C8B | 16163 | 8716 | 0.002 | -0.2626 | No | ||
| 54 | NEUROD2 | 20262 | 8856 | 0.002 | -0.2701 | No | ||
| 55 | TYRO3 | 5811 | 8858 | 0.002 | -0.2701 | No | ||
| 56 | CALD1 | 4273 8463 | 8892 | 0.001 | -0.2719 | No | ||
| 57 | LRRTM3 | 19744 | 9112 | 0.001 | -0.2837 | No | ||
| 58 | FOXA2 | 14405 2889 | 9330 | 0.000 | -0.2954 | No | ||
| 59 | GNAO1 | 4785 3829 | 9360 | 0.000 | -0.2970 | No | ||
| 60 | ALDH9A1 | 14064 | 9402 | 0.000 | -0.2992 | No | ||
| 61 | MAP3K13 | 22814 | 9607 | -0.000 | -0.3103 | No | ||
| 62 | ODZ1 | 24168 | 9659 | -0.000 | -0.3130 | No | ||
| 63 | TCF7L2 | 10048 5646 | 9715 | -0.001 | -0.3160 | No | ||
| 64 | EBF2 | 21975 | 9754 | -0.001 | -0.3180 | No | ||
| 65 | NRAS | 5191 | 10314 | -0.002 | -0.3482 | No | ||
| 66 | CYP7A1 | 15945 | 10507 | -0.003 | -0.3585 | No | ||
| 67 | PURA | 9670 | 10677 | -0.003 | -0.3676 | No | ||
| 68 | ITGA3 | 20284 | 10930 | -0.004 | -0.3811 | No | ||
| 69 | ANGPTL1 | 10195 8589 13064 | 11172 | -0.004 | -0.3941 | No | ||
| 70 | NR4A3 | 9473 16212 5183 | 11248 | -0.004 | -0.3980 | No | ||
| 71 | CHRDL1 | 8180 13504 | 11368 | -0.005 | -0.4043 | No | ||
| 72 | RGMA | 18213 | 11677 | -0.006 | -0.4208 | No | ||
| 73 | TJP1 | 17803 | 11707 | -0.006 | -0.4222 | No | ||
| 74 | PRDM1 | 19775 3337 | 11758 | -0.006 | -0.4248 | No | ||
| 75 | NFIA | 16172 5170 | 11867 | -0.006 | -0.4304 | No | ||
| 76 | FOXB1 | 12254 19069 | 11900 | -0.007 | -0.4320 | No | ||
| 77 | OTX2 | 9519 | 11914 | -0.007 | -0.4325 | No | ||
| 78 | BAMBI | 23629 | 11962 | -0.007 | -0.4349 | No | ||
| 79 | SPDEF | 23055 | 12502 | -0.009 | -0.4638 | No | ||
| 80 | PHEX | 24024 | 12526 | -0.009 | -0.4648 | No | ||
| 81 | HOXA11 | 17146 | 12539 | -0.009 | -0.4652 | No | ||
| 82 | HOXB8 | 9111 | 12624 | -0.010 | -0.4695 | No | ||
| 83 | PLAG1 | 7148 15949 12161 | 12679 | -0.010 | -0.4721 | No | ||
| 84 | PACSIN3 | 14948 8149 2776 2665 | 12700 | -0.010 | -0.4729 | No | ||
| 85 | ARHGEF2 | 4987 | 12818 | -0.011 | -0.4790 | No | ||
| 86 | TWIST2 | 14185 | 12997 | -0.012 | -0.4883 | No | ||
| 87 | LRP5 | 23948 9285 | 13023 | -0.012 | -0.4893 | No | ||
| 88 | VTN | 20756 | 13179 | -0.013 | -0.4974 | No | ||
| 89 | PLA2G1B | 9581 | 13366 | -0.015 | -0.5071 | No | ||
| 90 | FOXN1 | 20337 | 13484 | -0.015 | -0.5130 | No | ||
| 91 | FABP4 | 8601 | 13913 | -0.021 | -0.5356 | No | ||
| 92 | LCP2 | 4988 9268 | 14091 | -0.023 | -0.5446 | No | ||
| 93 | TLE1 | 5765 10185 15859 | 14152 | -0.024 | -0.5472 | No | ||
| 94 | NEO1 | 5161 | 14181 | -0.025 | -0.5480 | No | ||
| 95 | MGLL | 17368 1145 | 14183 | -0.025 | -0.5474 | No | ||
| 96 | CLDN2 | 4527 | 14207 | -0.025 | -0.5480 | No | ||
| 97 | C1QA | 8666 | 14306 | -0.027 | -0.5526 | No | ||
| 98 | FARP1 | 5849 10279 | 14428 | -0.029 | -0.5584 | No | ||
| 99 | TLE3 | 5767 19416 | 14511 | -0.031 | -0.5620 | No | ||
| 100 | GTF2A1 | 2064 8187 8188 13512 | 14773 | -0.038 | -0.5751 | No | ||
| 101 | GBX2 | 13887 | 14794 | -0.039 | -0.5751 | No | ||
| 102 | DMD | 24295 2647 | 14851 | -0.041 | -0.5771 | No | ||
| 103 | GNAZ | 71 | 14862 | -0.041 | -0.5765 | No | ||
| 104 | HAPLN1 | 21592 | 14916 | -0.044 | -0.5782 | No | ||
| 105 | SSH3 | 3709 10890 | 14976 | -0.046 | -0.5802 | No | ||
| 106 | FGF9 | 8966 | 15013 | -0.048 | -0.5808 | No | ||
| 107 | GPM6A | 3834 10618 | 15055 | -0.049 | -0.5817 | No | ||
| 108 | ITPKB | 3963 14028 | 15213 | -0.057 | -0.5887 | No | ||
| 109 | BAIAP2 | 449 4236 | 15285 | -0.061 | -0.5909 | No | ||
| 110 | BRUNOL4 | 2010 4229 | 15498 | -0.078 | -0.6003 | No | ||
| 111 | PDE4D | 10722 6235 | 15749 | -0.102 | -0.6111 | No | ||
| 112 | TGFB3 | 10161 | 15910 | -0.122 | -0.6165 | No | ||
| 113 | EGR2 | 8886 | 15957 | -0.132 | -0.6155 | No | ||
| 114 | MCC | 552 | 15993 | -0.138 | -0.6137 | No | ||
| 115 | PUM2 | 2071 8161 | 16197 | -0.184 | -0.6198 | No | ||
| 116 | PDCD4 | 5232 23816 | 16313 | -0.209 | -0.6204 | No | ||
| 117 | ZADH2 | 23501 | 16450 | -0.240 | -0.6214 | No | ||
| 118 | BUB3 | 18045 | 16538 | -0.267 | -0.6189 | No | ||
| 119 | PRKAG1 | 22135 | 17156 | -0.489 | -0.6393 | Yes | ||
| 120 | TOB1 | 20703 | 17217 | -0.513 | -0.6288 | Yes | ||
| 121 | AKT3 | 13739 982 | 17220 | -0.514 | -0.6152 | Yes | ||
| 122 | UHRF2 | 3731 4262 8452 23887 | 17296 | -0.545 | -0.6047 | Yes | ||
| 123 | LMO6 | 24392 | 17310 | -0.550 | -0.5907 | Yes | ||
| 124 | CHD2 | 10847 | 17320 | -0.556 | -0.5764 | Yes | ||
| 125 | GABARAPL1 | 17268 | 17405 | -0.598 | -0.5650 | Yes | ||
| 126 | PRDX5 | 7053 3732 | 17731 | -0.813 | -0.5608 | Yes | ||
| 127 | ZHX2 | 11842 | 17804 | -0.872 | -0.5415 | Yes | ||
| 128 | FOXP1 | 4242 | 17816 | -0.882 | -0.5185 | Yes | ||
| 129 | NDST2 | 21906 | 17830 | -0.889 | -0.4955 | Yes | ||
| 130 | LUC7L | 47 1520 | 17943 | -1.010 | -0.4746 | Yes | ||
| 131 | H2AFV | 1273 20531 | 17960 | -1.024 | -0.4481 | Yes | ||
| 132 | IL18 | 9172 | 18052 | -1.147 | -0.4224 | Yes | ||
| 133 | ABI1 | 4300 | 18144 | -1.265 | -0.3936 | Yes | ||
| 134 | UTX | 10266 2574 | 18165 | -1.301 | -0.3599 | Yes | ||
| 135 | PAFAH1B3 | 17928 | 18202 | -1.357 | -0.3256 | Yes | ||
| 136 | KLF3 | 4961 | 18251 | -1.482 | -0.2887 | Yes | ||
| 137 | CDC42EP3 | 6436 | 18383 | -1.819 | -0.2472 | Yes | ||
| 138 | CDKN1A | 4511 8729 | 18439 | -2.021 | -0.1962 | Yes | ||
| 139 | SGK | 9809 3419 | 18560 | -3.134 | -0.1191 | Yes | ||
| 140 | DUSP6 | 19891 3399 | 18597 | -4.571 | 0.0010 | Yes |

