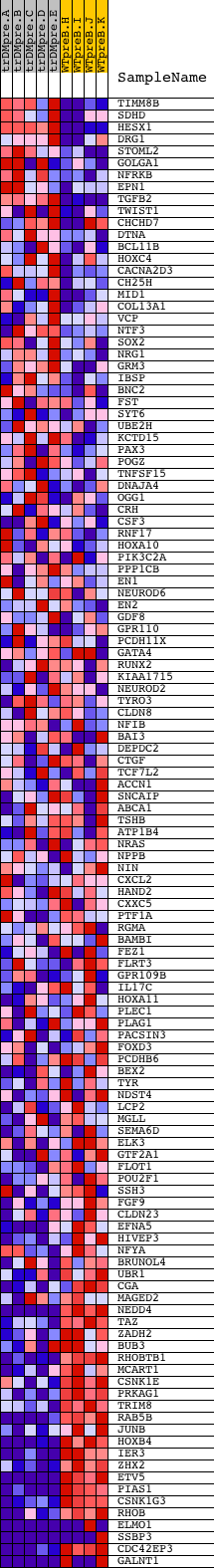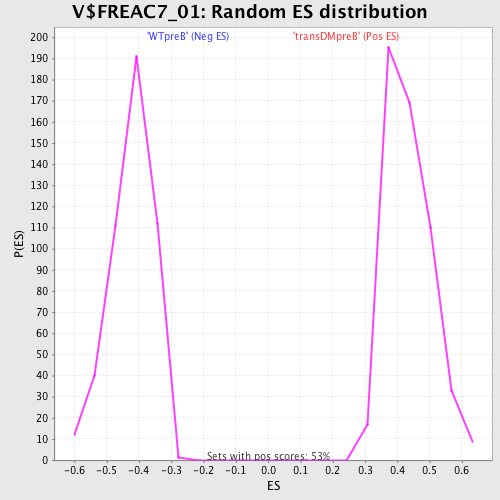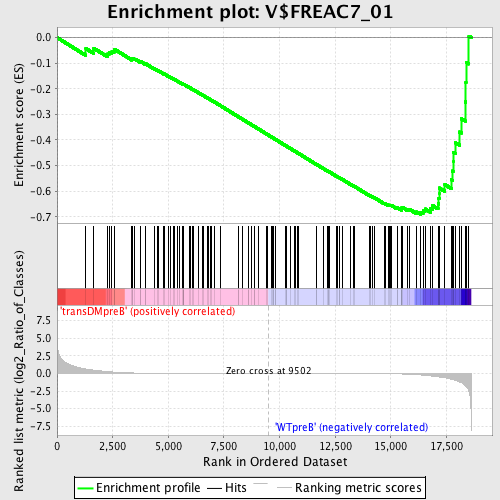
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$FREAC7_01 |
| Enrichment Score (ES) | -0.6911086 |
| Normalized Enrichment Score (NES) | -1.6334388 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0594513 |
| FWER p-Value | 0.141 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM8B | 11391 | 1293 | 0.625 | -0.0429 | No | ||
| 2 | SDHD | 19125 | 1650 | 0.470 | -0.0418 | No | ||
| 3 | HESX1 | 22070 | 2270 | 0.261 | -0.0640 | No | ||
| 4 | DRG1 | 1199 20553 | 2335 | 0.243 | -0.0570 | No | ||
| 5 | STOML2 | 15902 | 2465 | 0.207 | -0.0550 | No | ||
| 6 | GOLGA1 | 14595 | 2560 | 0.183 | -0.0522 | No | ||
| 7 | NFRKB | 10642 | 2587 | 0.175 | -0.0460 | No | ||
| 8 | EPN1 | 18401 | 3365 | 0.069 | -0.0850 | No | ||
| 9 | TGFB2 | 5743 | 3382 | 0.068 | -0.0830 | No | ||
| 10 | TWIST1 | 21291 | 3386 | 0.067 | -0.0802 | No | ||
| 11 | CHCHD7 | 16280 | 3497 | 0.060 | -0.0836 | No | ||
| 12 | DTNA | 8870 4647 23611 1991 | 3759 | 0.047 | -0.0957 | No | ||
| 13 | BCL11B | 7190 | 3768 | 0.046 | -0.0941 | No | ||
| 14 | HOXC4 | 4864 | 3955 | 0.039 | -0.1025 | No | ||
| 15 | CACNA2D3 | 8677 | 3972 | 0.039 | -0.1017 | No | ||
| 16 | CH25H | 23695 | 4356 | 0.028 | -0.1212 | No | ||
| 17 | MID1 | 5097 5098 | 4528 | 0.025 | -0.1293 | No | ||
| 18 | COL13A1 | 8763 | 4569 | 0.025 | -0.1304 | No | ||
| 19 | VCP | 11254 | 4796 | 0.021 | -0.1417 | No | ||
| 20 | NTF3 | 16991 | 4821 | 0.021 | -0.1421 | No | ||
| 21 | SOX2 | 9849 15612 | 4989 | 0.019 | -0.1503 | No | ||
| 22 | NRG1 | 3885 | 5109 | 0.017 | -0.1560 | No | ||
| 23 | GRM3 | 16929 | 5239 | 0.016 | -0.1623 | No | ||
| 24 | IBSP | 16775 | 5277 | 0.016 | -0.1636 | No | ||
| 25 | BNC2 | 15850 | 5416 | 0.015 | -0.1704 | No | ||
| 26 | FST | 8985 | 5505 | 0.014 | -0.1746 | No | ||
| 27 | SYT6 | 13437 7042 12040 | 5623 | 0.013 | -0.1803 | No | ||
| 28 | UBE2H | 5823 5822 | 5663 | 0.013 | -0.1819 | No | ||
| 29 | KCTD15 | 17864 | 5688 | 0.013 | -0.1826 | No | ||
| 30 | PAX3 | 13910 | 5690 | 0.013 | -0.1821 | No | ||
| 31 | POGZ | 6031 1911 | 5936 | 0.011 | -0.1949 | No | ||
| 32 | TNFSF15 | 15863 | 5973 | 0.011 | -0.1964 | No | ||
| 33 | DNAJA4 | 19446 | 6077 | 0.010 | -0.2015 | No | ||
| 34 | OGG1 | 17331 | 6135 | 0.010 | -0.2041 | No | ||
| 35 | CRH | 8784 | 6342 | 0.009 | -0.2149 | No | ||
| 36 | CSF3 | 1394 20671 | 6543 | 0.008 | -0.2253 | No | ||
| 37 | RNF17 | 11389 | 6561 | 0.008 | -0.2259 | No | ||
| 38 | HOXA10 | 17145 989 | 6759 | 0.008 | -0.2362 | No | ||
| 39 | PIK3C2A | 17667 | 6787 | 0.007 | -0.2373 | No | ||
| 40 | PPP1CB | 5282 3529 3504 | 6891 | 0.007 | -0.2426 | No | ||
| 41 | EN1 | 387 14155 | 6944 | 0.007 | -0.2451 | No | ||
| 42 | NEUROD6 | 17138 | 7095 | 0.006 | -0.2530 | No | ||
| 43 | EN2 | 16898 | 7358 | 0.006 | -0.2669 | No | ||
| 44 | GDF8 | 9411 | 8130 | 0.003 | -0.3084 | No | ||
| 45 | GPR110 | 23234 | 8164 | 0.003 | -0.3101 | No | ||
| 46 | PCDH11X | 2559 | 8349 | 0.003 | -0.3199 | No | ||
| 47 | GATA4 | 21792 4755 | 8608 | 0.002 | -0.3337 | No | ||
| 48 | RUNX2 | 4480 8700 | 8624 | 0.002 | -0.3345 | No | ||
| 49 | KIAA1715 | 7629 | 8738 | 0.002 | -0.3405 | No | ||
| 50 | NEUROD2 | 20262 | 8856 | 0.002 | -0.3468 | No | ||
| 51 | TYRO3 | 5811 | 8858 | 0.002 | -0.3467 | No | ||
| 52 | CLDN8 | 22549 1747 | 9068 | 0.001 | -0.3580 | No | ||
| 53 | NFIB | 15855 | 9395 | 0.000 | -0.3756 | No | ||
| 54 | BAI3 | 5580 | 9437 | 0.000 | -0.3778 | No | ||
| 55 | DEPDC2 | 14295 | 9655 | -0.000 | -0.3895 | No | ||
| 56 | CTGF | 20068 | 9676 | -0.000 | -0.3906 | No | ||
| 57 | TCF7L2 | 10048 5646 | 9715 | -0.001 | -0.3926 | No | ||
| 58 | ACCN1 | 20327 1194 | 9717 | -0.001 | -0.3927 | No | ||
| 59 | SNCAIP | 12589 | 9829 | -0.001 | -0.3986 | No | ||
| 60 | ABCA1 | 15881 | 9836 | -0.001 | -0.3989 | No | ||
| 61 | TSHB | 15218 | 10280 | -0.002 | -0.4228 | No | ||
| 62 | ATP1B4 | 12586 | 10310 | -0.002 | -0.4243 | No | ||
| 63 | NRAS | 5191 | 10314 | -0.002 | -0.4243 | No | ||
| 64 | NPPB | 15994 | 10485 | -0.002 | -0.4334 | No | ||
| 65 | NIN | 9463 2103 | 10679 | -0.003 | -0.4437 | No | ||
| 66 | CXCL2 | 9790 | 10702 | -0.003 | -0.4448 | No | ||
| 67 | HAND2 | 18615 | 10785 | -0.003 | -0.4491 | No | ||
| 68 | CXXC5 | 12523 | 10860 | -0.003 | -0.4529 | No | ||
| 69 | PTF1A | 15110 | 11667 | -0.006 | -0.4963 | No | ||
| 70 | RGMA | 18213 | 11677 | -0.006 | -0.4965 | No | ||
| 71 | BAMBI | 23629 | 11962 | -0.007 | -0.5115 | No | ||
| 72 | FEZ1 | 19520 3100 | 12150 | -0.008 | -0.5213 | No | ||
| 73 | FLRT3 | 7730 12930 2673 | 12177 | -0.008 | -0.5224 | No | ||
| 74 | GPR109B | 16374 | 12210 | -0.008 | -0.5238 | No | ||
| 75 | IL17C | 18438 | 12258 | -0.008 | -0.5260 | No | ||
| 76 | HOXA11 | 17146 | 12539 | -0.009 | -0.5407 | No | ||
| 77 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 12582 | -0.010 | -0.5426 | No | ||
| 78 | PLAG1 | 7148 15949 12161 | 12679 | -0.010 | -0.5473 | No | ||
| 79 | PACSIN3 | 14948 8149 2776 2665 | 12700 | -0.010 | -0.5480 | No | ||
| 80 | FOXD3 | 9088 | 12809 | -0.011 | -0.5534 | No | ||
| 81 | PCDHB6 | 8224 | 13184 | -0.013 | -0.5730 | No | ||
| 82 | BEX2 | 24055 | 13341 | -0.014 | -0.5808 | No | ||
| 83 | TYR | 17763 | 13346 | -0.014 | -0.5804 | No | ||
| 84 | NDST4 | 12259 7228 | 14028 | -0.022 | -0.6163 | No | ||
| 85 | LCP2 | 4988 9268 | 14091 | -0.023 | -0.6186 | No | ||
| 86 | MGLL | 17368 1145 | 14183 | -0.025 | -0.6225 | No | ||
| 87 | SEMA6D | 2800 14876 90 | 14253 | -0.026 | -0.6251 | No | ||
| 88 | ELK3 | 4663 94 3388 | 14711 | -0.037 | -0.6483 | No | ||
| 89 | GTF2A1 | 2064 8187 8188 13512 | 14773 | -0.038 | -0.6499 | No | ||
| 90 | FLOT1 | 23249 1613 | 14894 | -0.043 | -0.6545 | No | ||
| 91 | POU2F1 | 5275 3989 4065 4010 | 14921 | -0.044 | -0.6541 | No | ||
| 92 | SSH3 | 3709 10890 | 14976 | -0.046 | -0.6550 | No | ||
| 93 | FGF9 | 8966 | 15013 | -0.048 | -0.6549 | No | ||
| 94 | CLDN23 | 18893 | 15282 | -0.061 | -0.6667 | No | ||
| 95 | EFNA5 | 4655 8884 | 15286 | -0.061 | -0.6643 | No | ||
| 96 | HIVEP3 | 4973 | 15480 | -0.076 | -0.6714 | No | ||
| 97 | NFYA | 5172 | 15486 | -0.077 | -0.6683 | No | ||
| 98 | BRUNOL4 | 2010 4229 | 15498 | -0.078 | -0.6655 | No | ||
| 99 | UBR1 | 5828 | 15523 | -0.080 | -0.6634 | No | ||
| 100 | CGA | 16246 | 15768 | -0.103 | -0.6721 | No | ||
| 101 | MAGED2 | 24034 | 15859 | -0.114 | -0.6721 | No | ||
| 102 | NEDD4 | 19384 3101 | 16169 | -0.177 | -0.6811 | No | ||
| 103 | TAZ | 12415 | 16355 | -0.219 | -0.6816 | Yes | ||
| 104 | ZADH2 | 23501 | 16450 | -0.240 | -0.6763 | Yes | ||
| 105 | BUB3 | 18045 | 16538 | -0.267 | -0.6695 | Yes | ||
| 106 | RHOBTB1 | 12789 | 16786 | -0.344 | -0.6680 | Yes | ||
| 107 | MCART1 | 10492 | 16873 | -0.384 | -0.6560 | Yes | ||
| 108 | CSNK1E | 6570 2211 11332 | 17125 | -0.478 | -0.6489 | Yes | ||
| 109 | PRKAG1 | 22135 | 17156 | -0.489 | -0.6294 | Yes | ||
| 110 | TRIM8 | 23824 | 17173 | -0.497 | -0.6088 | Yes | ||
| 111 | RAB5B | 359 19591 3430 | 17180 | -0.501 | -0.5874 | Yes | ||
| 112 | JUNB | 9201 | 17428 | -0.615 | -0.5742 | Yes | ||
| 113 | HOXB4 | 20686 | 17727 | -0.811 | -0.5553 | Yes | ||
| 114 | IER3 | 23248 | 17765 | -0.839 | -0.5210 | Yes | ||
| 115 | ZHX2 | 11842 | 17804 | -0.872 | -0.4854 | Yes | ||
| 116 | ETV5 | 22630 | 17811 | -0.879 | -0.4477 | Yes | ||
| 117 | PIAS1 | 7126 | 17914 | -0.971 | -0.4112 | Yes | ||
| 118 | CSNK1G3 | 12873 | 18086 | -1.197 | -0.3687 | Yes | ||
| 119 | RHOB | 4411 | 18166 | -1.305 | -0.3166 | Yes | ||
| 120 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 18347 | -1.737 | -0.2512 | Yes | ||
| 121 | SSBP3 | 2439 7814 2431 | 18373 | -1.803 | -0.1746 | Yes | ||
| 122 | CDC42EP3 | 6436 | 18383 | -1.819 | -0.0965 | Yes | ||
| 123 | GALNT1 | 8998 23609 8999 4750 | 18510 | -2.522 | 0.0057 | Yes |