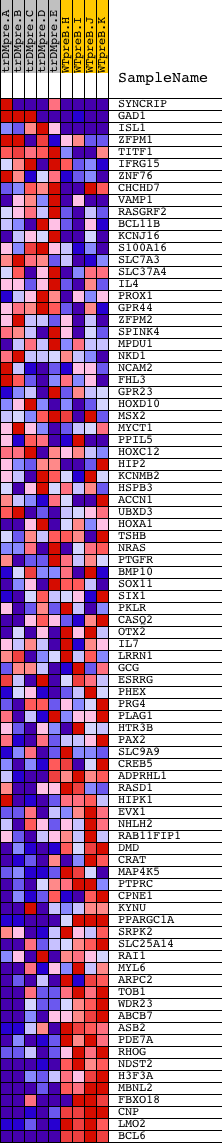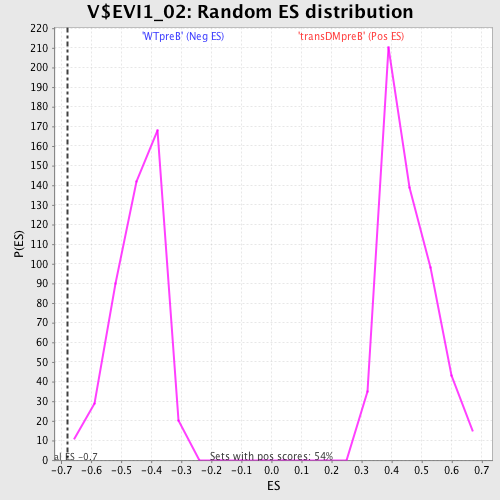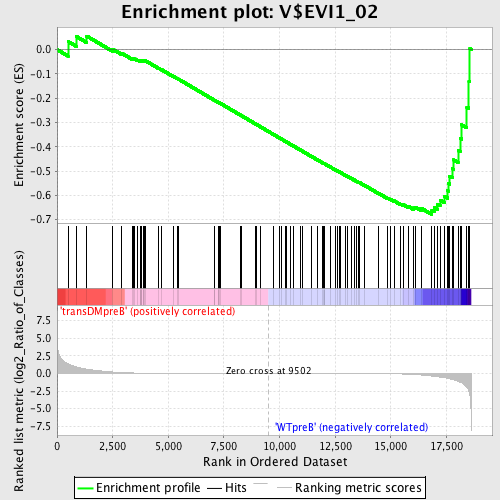
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$EVI1_02 |
| Enrichment Score (ES) | -0.6786649 |
| Normalized Enrichment Score (NES) | -1.5288832 |
| Nominal p-value | 0.006521739 |
| FDR q-value | 0.15062243 |
| FWER p-Value | 0.646 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SYNCRIP | 3078 3035 3107 | 496 | 1.355 | 0.0334 | No | ||
| 2 | GAD1 | 14993 2690 | 876 | 0.913 | 0.0534 | No | ||
| 3 | ISL1 | 21338 | 1334 | 0.604 | 0.0555 | No | ||
| 4 | ZFPM1 | 18439 | 2490 | 0.199 | 0.0020 | No | ||
| 5 | TITF1 | 10180 21062 | 2912 | 0.120 | -0.0154 | No | ||
| 6 | IFRG15 | 7222 | 3407 | 0.065 | -0.0391 | No | ||
| 7 | ZNF76 | 23320 | 3445 | 0.063 | -0.0383 | No | ||
| 8 | CHCHD7 | 16280 | 3497 | 0.060 | -0.0384 | No | ||
| 9 | VAMP1 | 5846 1043 | 3625 | 0.053 | -0.0430 | No | ||
| 10 | RASGRF2 | 21397 | 3730 | 0.048 | -0.0464 | No | ||
| 11 | BCL11B | 7190 | 3768 | 0.046 | -0.0464 | No | ||
| 12 | KCNJ16 | 20613 | 3783 | 0.046 | -0.0451 | No | ||
| 13 | S100A16 | 15530 | 3802 | 0.045 | -0.0441 | No | ||
| 14 | SLC7A3 | 2608 24097 | 3871 | 0.042 | -0.0459 | No | ||
| 15 | SLC37A4 | 8994 | 3879 | 0.042 | -0.0444 | No | ||
| 16 | IL4 | 9174 | 3933 | 0.040 | -0.0455 | No | ||
| 17 | PROX1 | 9623 | 3979 | 0.039 | -0.0462 | No | ||
| 18 | GPR44 | 23926 | 4548 | 0.025 | -0.0757 | No | ||
| 19 | ZFPM2 | 22483 | 4704 | 0.022 | -0.0831 | No | ||
| 20 | SPINK4 | 16241 | 5214 | 0.016 | -0.1098 | No | ||
| 21 | MPDU1 | 20386 | 5397 | 0.015 | -0.1190 | No | ||
| 22 | NKD1 | 8228 | 5474 | 0.014 | -0.1225 | No | ||
| 23 | NCAM2 | 9445 5150 | 7064 | 0.007 | -0.2079 | No | ||
| 24 | FHL3 | 16092 2457 | 7084 | 0.006 | -0.2087 | No | ||
| 25 | GPR23 | 13428 | 7237 | 0.006 | -0.2166 | No | ||
| 26 | HOXD10 | 14983 | 7260 | 0.006 | -0.2176 | No | ||
| 27 | MSX2 | 21461 | 7302 | 0.006 | -0.2195 | No | ||
| 28 | MYCT1 | 7572 | 7335 | 0.006 | -0.2210 | No | ||
| 29 | PPIL5 | 21255 | 8251 | 0.003 | -0.2702 | No | ||
| 30 | HOXC12 | 22343 | 8309 | 0.003 | -0.2732 | No | ||
| 31 | HIP2 | 3626 3485 16838 | 8926 | 0.001 | -0.3064 | No | ||
| 32 | KCNMB2 | 1939 15615 | 8955 | 0.001 | -0.3078 | No | ||
| 33 | HSPB3 | 21340 | 9153 | 0.001 | -0.3184 | No | ||
| 34 | ACCN1 | 20327 1194 | 9717 | -0.001 | -0.3488 | No | ||
| 35 | UBXD3 | 15701 | 10015 | -0.001 | -0.3647 | No | ||
| 36 | HOXA1 | 1005 17152 | 10103 | -0.002 | -0.3694 | No | ||
| 37 | TSHB | 15218 | 10280 | -0.002 | -0.3788 | No | ||
| 38 | NRAS | 5191 | 10314 | -0.002 | -0.3805 | No | ||
| 39 | PTGFR | 15138 | 10503 | -0.003 | -0.3905 | No | ||
| 40 | BMP10 | 17376 | 10617 | -0.003 | -0.3965 | No | ||
| 41 | SOX11 | 5477 | 10943 | -0.004 | -0.4138 | No | ||
| 42 | SIX1 | 9821 | 11012 | -0.004 | -0.4173 | No | ||
| 43 | PKLR | 1850 15545 | 11427 | -0.005 | -0.4395 | No | ||
| 44 | CASQ2 | 15477 | 11683 | -0.006 | -0.4530 | No | ||
| 45 | OTX2 | 9519 | 11914 | -0.007 | -0.4651 | No | ||
| 46 | IL7 | 4921 | 11981 | -0.007 | -0.4683 | No | ||
| 47 | LRRN1 | 17343 | 12007 | -0.007 | -0.4694 | No | ||
| 48 | GCG | 14578 | 12016 | -0.007 | -0.4695 | No | ||
| 49 | ESRRG | 6447 | 12271 | -0.008 | -0.4828 | No | ||
| 50 | PHEX | 24024 | 12526 | -0.009 | -0.4961 | No | ||
| 51 | PRG4 | 13592 4036 | 12600 | -0.010 | -0.4996 | No | ||
| 52 | PLAG1 | 7148 15949 12161 | 12679 | -0.010 | -0.5034 | No | ||
| 53 | HTR3B | 19129 3145 | 12726 | -0.010 | -0.5054 | No | ||
| 54 | PAX2 | 9530 24416 | 12956 | -0.012 | -0.5173 | No | ||
| 55 | SLC9A9 | 11661 | 13041 | -0.012 | -0.5213 | No | ||
| 56 | CREB5 | 10551 | 13229 | -0.013 | -0.5307 | No | ||
| 57 | ADPRHL1 | 18930 3863 | 13345 | -0.014 | -0.5363 | No | ||
| 58 | RASD1 | 20424 | 13467 | -0.015 | -0.5422 | No | ||
| 59 | HIPK1 | 4851 | 13537 | -0.016 | -0.5452 | No | ||
| 60 | EVX1 | 17442 | 13574 | -0.016 | -0.5464 | No | ||
| 61 | NHLH2 | 9462 | 13802 | -0.019 | -0.5578 | No | ||
| 62 | RAB11FIP1 | 13287 | 14446 | -0.030 | -0.5912 | No | ||
| 63 | DMD | 24295 2647 | 14851 | -0.041 | -0.6112 | No | ||
| 64 | CRAT | 4557 2804 2896 | 14979 | -0.046 | -0.6160 | No | ||
| 65 | MAP4K5 | 11853 21048 2101 2142 | 15149 | -0.053 | -0.6227 | No | ||
| 66 | PTPRC | 5327 9662 | 15455 | -0.074 | -0.6359 | No | ||
| 67 | CPNE1 | 11186 | 15562 | -0.083 | -0.6380 | No | ||
| 68 | KYNU | 15016 | 15772 | -0.103 | -0.6447 | No | ||
| 69 | PPARGC1A | 16533 | 16024 | -0.143 | -0.6518 | No | ||
| 70 | SRPK2 | 5513 | 16098 | -0.163 | -0.6486 | No | ||
| 71 | SLC25A14 | 24344 | 16386 | -0.226 | -0.6540 | No | ||
| 72 | RAI1 | 5355 20861 | 16844 | -0.370 | -0.6622 | Yes | ||
| 73 | MYL6 | 9438 3408 | 16952 | -0.412 | -0.6497 | Yes | ||
| 74 | ARPC2 | 14225 | 17100 | -0.470 | -0.6368 | Yes | ||
| 75 | TOB1 | 20703 | 17217 | -0.513 | -0.6203 | Yes | ||
| 76 | WDR23 | 22009 | 17402 | -0.598 | -0.6037 | Yes | ||
| 77 | ABCB7 | 24085 | 17557 | -0.692 | -0.5813 | Yes | ||
| 78 | ASB2 | 20996 | 17598 | -0.714 | -0.5517 | Yes | ||
| 79 | PDE7A | 9544 1788 | 17646 | -0.750 | -0.5210 | Yes | ||
| 80 | RHOG | 17727 | 17770 | -0.843 | -0.4902 | Yes | ||
| 81 | NDST2 | 21906 | 17830 | -0.889 | -0.4540 | Yes | ||
| 82 | H3F3A | 4838 | 18026 | -1.107 | -0.4154 | Yes | ||
| 83 | MBNL2 | 21932 | 18117 | -1.242 | -0.3651 | Yes | ||
| 84 | FBXO18 | 14686 2796 | 18186 | -1.332 | -0.3097 | Yes | ||
| 85 | CNP | 8760 | 18419 | -1.914 | -0.2373 | Yes | ||
| 86 | LMO2 | 9281 | 18511 | -2.525 | -0.1302 | Yes | ||
| 87 | BCL6 | 22624 | 18554 | -3.060 | 0.0033 | Yes |