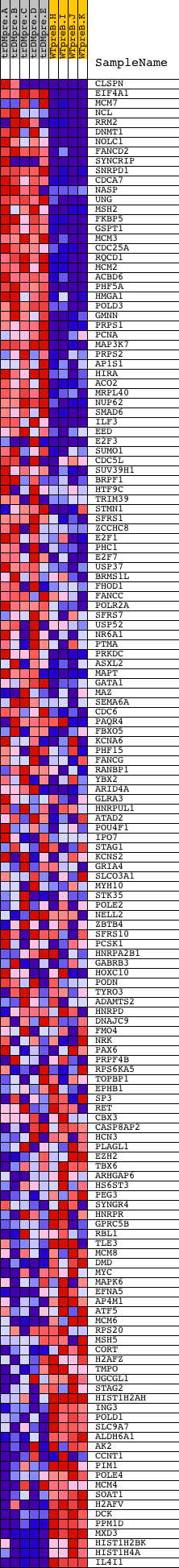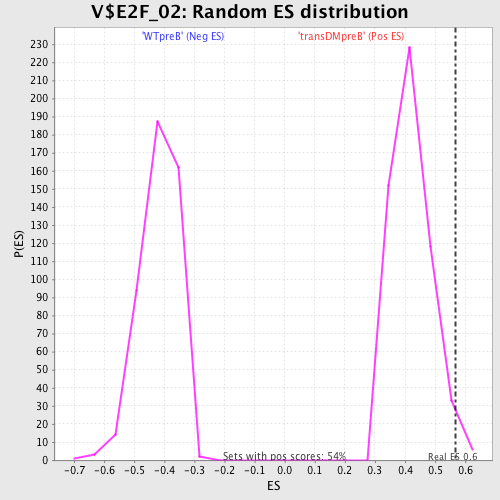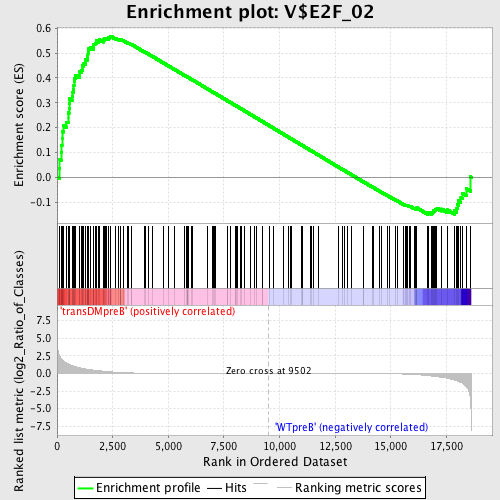
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | V$E2F_02 |
| Enrichment Score (ES) | 0.56863976 |
| Normalized Enrichment Score (NES) | 1.3457674 |
| Nominal p-value | 0.016759777 |
| FDR q-value | 0.5571349 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CLSPN | 16079 11256 6530 | 94 | 2.627 | 0.0349 | Yes | ||
| 2 | EIF4A1 | 8889 23719 | 122 | 2.399 | 0.0699 | Yes | ||
| 3 | MCM7 | 9372 3568 | 178 | 2.099 | 0.0989 | Yes | ||
| 4 | NCL | 5153 13899 | 191 | 2.049 | 0.1294 | Yes | ||
| 5 | RRM2 | 5401 5400 | 222 | 1.924 | 0.1570 | Yes | ||
| 6 | DNMT1 | 19217 | 226 | 1.914 | 0.1860 | Yes | ||
| 7 | NOLC1 | 7704 | 300 | 1.658 | 0.2073 | Yes | ||
| 8 | FANCD2 | 17326 464 | 435 | 1.463 | 0.2223 | Yes | ||
| 9 | SYNCRIP | 3078 3035 3107 | 496 | 1.355 | 0.2396 | Yes | ||
| 10 | SNRPD1 | 23622 | 497 | 1.354 | 0.2602 | Yes | ||
| 11 | CDCA7 | 12437 | 536 | 1.297 | 0.2779 | Yes | ||
| 12 | NASP | 2383 2387 6955 | 545 | 1.283 | 0.2970 | Yes | ||
| 13 | UNG | 10257 16744 | 556 | 1.255 | 0.3155 | Yes | ||
| 14 | MSH2 | 23138 | 676 | 1.101 | 0.3258 | Yes | ||
| 15 | FKBP5 | 23051 | 693 | 1.087 | 0.3415 | Yes | ||
| 16 | GSPT1 | 9046 | 730 | 1.047 | 0.3555 | Yes | ||
| 17 | MCM3 | 13991 | 755 | 1.026 | 0.3698 | Yes | ||
| 18 | CDC25A | 8721 | 763 | 1.018 | 0.3849 | Yes | ||
| 19 | RQCD1 | 12205 | 782 | 0.999 | 0.3991 | Yes | ||
| 20 | MCM2 | 17074 | 826 | 0.958 | 0.4114 | Yes | ||
| 21 | ACBD6 | 14089 | 1019 | 0.792 | 0.4130 | Yes | ||
| 22 | PHF5A | 22194 | 1022 | 0.790 | 0.4249 | Yes | ||
| 23 | HMGA1 | 23323 | 1108 | 0.730 | 0.4314 | Yes | ||
| 24 | POLD3 | 17742 | 1142 | 0.708 | 0.4404 | Yes | ||
| 25 | GMNN | 21513 | 1145 | 0.706 | 0.4511 | Yes | ||
| 26 | PRPS1 | 24233 | 1205 | 0.671 | 0.4581 | Yes | ||
| 27 | PCNA | 9535 | 1278 | 0.629 | 0.4638 | Yes | ||
| 28 | MAP3K7 | 16255 | 1281 | 0.628 | 0.4732 | Yes | ||
| 29 | PRPS2 | 24003 | 1351 | 0.594 | 0.4785 | Yes | ||
| 30 | AP1S1 | 3500 3453 16335 | 1382 | 0.578 | 0.4857 | Yes | ||
| 31 | HIRA | 4852 9090 | 1386 | 0.577 | 0.4943 | Yes | ||
| 32 | ACO2 | 8527 | 1389 | 0.575 | 0.5029 | Yes | ||
| 33 | MRPL40 | 22641 | 1397 | 0.571 | 0.5112 | Yes | ||
| 34 | NUP62 | 9497 | 1427 | 0.558 | 0.5182 | Yes | ||
| 35 | SMAD6 | 19083 | 1493 | 0.533 | 0.5227 | Yes | ||
| 36 | ILF3 | 3110 3030 9176 | 1627 | 0.481 | 0.5229 | Yes | ||
| 37 | EED | 17759 | 1632 | 0.478 | 0.5299 | Yes | ||
| 38 | E2F3 | 21500 8874 | 1639 | 0.476 | 0.5368 | Yes | ||
| 39 | SUMO1 | 5826 3943 10247 | 1708 | 0.452 | 0.5400 | Yes | ||
| 40 | CDC5L | 7746 7745 7744 | 1766 | 0.431 | 0.5435 | Yes | ||
| 41 | SUV39H1 | 24194 | 1774 | 0.430 | 0.5497 | Yes | ||
| 42 | BRPF1 | 17332 | 1872 | 0.385 | 0.5503 | Yes | ||
| 43 | HTF9C | 4886 1678 | 1908 | 0.374 | 0.5541 | Yes | ||
| 44 | TRIM39 | 22994 | 2080 | 0.319 | 0.5497 | Yes | ||
| 45 | STMN1 | 9261 | 2099 | 0.313 | 0.5534 | Yes | ||
| 46 | SFRS1 | 8492 | 2114 | 0.308 | 0.5574 | Yes | ||
| 47 | ZCCHC8 | 3570 7696 | 2176 | 0.290 | 0.5585 | Yes | ||
| 48 | E2F1 | 14384 | 2208 | 0.278 | 0.5610 | Yes | ||
| 49 | PHC1 | 17013 | 2312 | 0.251 | 0.5593 | Yes | ||
| 50 | E2F7 | 19884 | 2314 | 0.251 | 0.5630 | Yes | ||
| 51 | USP37 | 13923 | 2377 | 0.233 | 0.5632 | Yes | ||
| 52 | BRMS1L | 21268 | 2399 | 0.227 | 0.5655 | Yes | ||
| 53 | FHOD1 | 18772 | 2406 | 0.226 | 0.5686 | Yes | ||
| 54 | FANCC | 4712 | 2628 | 0.168 | 0.5592 | No | ||
| 55 | POLR2A | 5394 | 2747 | 0.149 | 0.5551 | No | ||
| 56 | SFRS7 | 22889 | 2839 | 0.130 | 0.5522 | No | ||
| 57 | USP52 | 3397 19841 | 2840 | 0.130 | 0.5541 | No | ||
| 58 | NR6A1 | 14596 | 2857 | 0.128 | 0.5552 | No | ||
| 59 | PTMA | 9657 | 2964 | 0.111 | 0.5512 | No | ||
| 60 | PRKDC | 9617 22847 | 3163 | 0.086 | 0.5418 | No | ||
| 61 | ASXL2 | 7957 | 3226 | 0.080 | 0.5396 | No | ||
| 62 | MAPT | 9420 1347 | 3342 | 0.071 | 0.5345 | No | ||
| 63 | GATA1 | 24196 | 3343 | 0.071 | 0.5355 | No | ||
| 64 | MAZ | 1327 17623 | 3923 | 0.040 | 0.5048 | No | ||
| 65 | SEMA6A | 23439 9802 | 3967 | 0.039 | 0.5031 | No | ||
| 66 | CDC6 | 6221 6220 | 4092 | 0.035 | 0.4969 | No | ||
| 67 | PAQR4 | 23105 | 4294 | 0.030 | 0.4864 | No | ||
| 68 | FBXO5 | 19830 | 4790 | 0.021 | 0.4600 | No | ||
| 69 | KCNA6 | 664 16990 665 4938 | 5017 | 0.019 | 0.4480 | No | ||
| 70 | PHF15 | 20467 8050 | 5286 | 0.016 | 0.4337 | No | ||
| 71 | FANCG | 15904 | 5739 | 0.012 | 0.4094 | No | ||
| 72 | RANBP1 | 9692 5357 | 5795 | 0.012 | 0.4066 | No | ||
| 73 | YBX2 | 20818 | 5850 | 0.012 | 0.4039 | No | ||
| 74 | ARID4A | 2080 6215 | 5859 | 0.012 | 0.4036 | No | ||
| 75 | GLRA3 | 18617 | 5871 | 0.012 | 0.4032 | No | ||
| 76 | HNRPUL1 | 6118 17923 1608 10577 1345 | 5900 | 0.011 | 0.4019 | No | ||
| 77 | ATAD2 | 2268 2256 | 6043 | 0.011 | 0.3943 | No | ||
| 78 | POU4F1 | 21727 | 6064 | 0.011 | 0.3934 | No | ||
| 79 | IPO7 | 6130 | 6752 | 0.008 | 0.3563 | No | ||
| 80 | STAG1 | 5520 9905 2987 | 7004 | 0.007 | 0.3428 | No | ||
| 81 | KCNS2 | 4949 2232 | 7008 | 0.007 | 0.3428 | No | ||
| 82 | GRIA4 | 19249 | 7080 | 0.006 | 0.3390 | No | ||
| 83 | SLCO3A1 | 17796 4237 8430 | 7124 | 0.006 | 0.3368 | No | ||
| 84 | MYH10 | 8077 | 7678 | 0.005 | 0.3069 | No | ||
| 85 | STK35 | 14851 | 7679 | 0.005 | 0.3070 | No | ||
| 86 | POLE2 | 21053 | 7815 | 0.004 | 0.2997 | No | ||
| 87 | NELL2 | 12006 7020 22151 | 7997 | 0.004 | 0.2900 | No | ||
| 88 | ZBTB4 | 13267 | 8020 | 0.004 | 0.2888 | No | ||
| 89 | SFRS10 | 9818 1722 5441 | 8050 | 0.004 | 0.2873 | No | ||
| 90 | PCSK1 | 21600 | 8126 | 0.003 | 0.2833 | No | ||
| 91 | HNRPA2B1 | 1187 7001 7000 1040 | 8229 | 0.003 | 0.2778 | No | ||
| 92 | GABRB3 | 4747 | 8279 | 0.003 | 0.2752 | No | ||
| 93 | HOXC10 | 22342 | 8431 | 0.003 | 0.2671 | No | ||
| 94 | PODN | 15811 | 8681 | 0.002 | 0.2536 | No | ||
| 95 | TYRO3 | 5811 | 8858 | 0.002 | 0.2441 | No | ||
| 96 | ADAMTS2 | 5706 20899 1356 | 8974 | 0.001 | 0.2379 | No | ||
| 97 | HNRPD | 8645 | 9217 | 0.001 | 0.2248 | No | ||
| 98 | DNAJC9 | 21912 | 9225 | 0.001 | 0.2245 | No | ||
| 99 | FMO4 | 13786 | 9537 | -0.000 | 0.2076 | No | ||
| 100 | NRK | 6559 | 9713 | -0.001 | 0.1981 | No | ||
| 101 | PAX6 | 5223 | 10181 | -0.002 | 0.1729 | No | ||
| 102 | PRPF4B | 3208 5295 | 10398 | -0.002 | 0.1612 | No | ||
| 103 | RPS6KA5 | 2076 21005 | 10489 | -0.002 | 0.1564 | No | ||
| 104 | TOPBP1 | 10659 | 10535 | -0.003 | 0.1540 | No | ||
| 105 | EPHB1 | 19024 | 10967 | -0.004 | 0.1307 | No | ||
| 106 | SP3 | 5483 | 11020 | -0.004 | 0.1279 | No | ||
| 107 | RET | 17028 | 11047 | -0.004 | 0.1266 | No | ||
| 108 | CBX3 | 8705 4484 | 11398 | -0.005 | 0.1077 | No | ||
| 109 | CASP8AP2 | 16253 | 11426 | -0.005 | 0.1063 | No | ||
| 110 | HCN3 | 4843 1763 | 11506 | -0.005 | 0.1021 | No | ||
| 111 | PLAGL1 | 10372 3414 | 11760 | -0.006 | 0.0885 | No | ||
| 112 | EZH2 | 1092 17163 | 12637 | -0.010 | 0.0412 | No | ||
| 113 | TBX6 | 1993 18075 | 12820 | -0.011 | 0.0315 | No | ||
| 114 | ARHGAP6 | 24207 | 12924 | -0.011 | 0.0261 | No | ||
| 115 | HS6ST3 | 21933 | 13035 | -0.012 | 0.0203 | No | ||
| 116 | PEG3 | 5240 | 13221 | -0.013 | 0.0105 | No | ||
| 117 | SYNGR4 | 17825 | 13761 | -0.018 | -0.0184 | No | ||
| 118 | HNRPR | 13196 7909 2500 | 13777 | -0.019 | -0.0189 | No | ||
| 119 | GPRC5B | 17662 | 14171 | -0.024 | -0.0398 | No | ||
| 120 | RBL1 | 14373 | 14215 | -0.025 | -0.0418 | No | ||
| 121 | TLE3 | 5767 19416 | 14511 | -0.031 | -0.0573 | No | ||
| 122 | MCM8 | 14834 | 14573 | -0.033 | -0.0601 | No | ||
| 123 | DMD | 24295 2647 | 14851 | -0.041 | -0.0745 | No | ||
| 124 | MYC | 22465 9435 | 14956 | -0.045 | -0.0794 | No | ||
| 125 | MAPK6 | 19062 | 15200 | -0.056 | -0.0917 | No | ||
| 126 | EFNA5 | 4655 8884 | 15286 | -0.061 | -0.0954 | No | ||
| 127 | AP4M1 | 3499 16656 | 15591 | -0.085 | -0.1106 | No | ||
| 128 | ATF5 | 17843 | 15646 | -0.090 | -0.1121 | No | ||
| 129 | MCM6 | 4000 13845 4119 | 15695 | -0.095 | -0.1133 | No | ||
| 130 | RPS20 | 7438 | 15729 | -0.098 | -0.1136 | No | ||
| 131 | MSH5 | 23010 1599 | 15846 | -0.112 | -0.1182 | No | ||
| 132 | CORT | 15669 2539 | 15867 | -0.115 | -0.1175 | No | ||
| 133 | H2AFZ | 6957 | 16059 | -0.151 | -0.1255 | No | ||
| 134 | TMPO | 5778 | 16117 | -0.167 | -0.1261 | No | ||
| 135 | UGCGL1 | 6721 | 16136 | -0.170 | -0.1245 | No | ||
| 136 | STAG2 | 5521 | 16171 | -0.178 | -0.1236 | No | ||
| 137 | HIST1H2AH | 11414 | 16657 | -0.302 | -0.1453 | No | ||
| 138 | ING3 | 17514 1019 | 16687 | -0.312 | -0.1421 | No | ||
| 139 | POLD1 | 17847 | 16839 | -0.368 | -0.1447 | No | ||
| 140 | SLC9A7 | 24177 | 16886 | -0.389 | -0.1413 | No | ||
| 141 | ALDH6A1 | 21025 | 16900 | -0.394 | -0.1360 | No | ||
| 142 | AK2 | 8563 2479 16073 | 16960 | -0.419 | -0.1328 | No | ||
| 143 | CCNT1 | 22140 11607 | 17006 | -0.435 | -0.1286 | No | ||
| 144 | PIM1 | 23308 | 17073 | -0.461 | -0.1252 | No | ||
| 145 | POLE4 | 12441 | 17279 | -0.536 | -0.1281 | No | ||
| 146 | MCM4 | 22655 1708 | 17543 | -0.687 | -0.1319 | No | ||
| 147 | SOAT1 | 13797 | 17857 | -0.911 | -0.1350 | No | ||
| 148 | H2AFV | 1273 20531 | 17960 | -1.024 | -0.1250 | No | ||
| 149 | DCK | 16808 | 17984 | -1.054 | -0.1102 | No | ||
| 150 | PPM1D | 20721 | 18030 | -1.116 | -0.0957 | No | ||
| 151 | MXD3 | 21455 | 18151 | -1.270 | -0.0828 | No | ||
| 152 | HIST1H2BK | 11423 | 18232 | -1.432 | -0.0654 | No | ||
| 153 | HIST1H4A | 11590 | 18396 | -1.845 | -0.0461 | No | ||
| 154 | IL4I1 | 8971 | 18580 | -3.813 | 0.0020 | No |