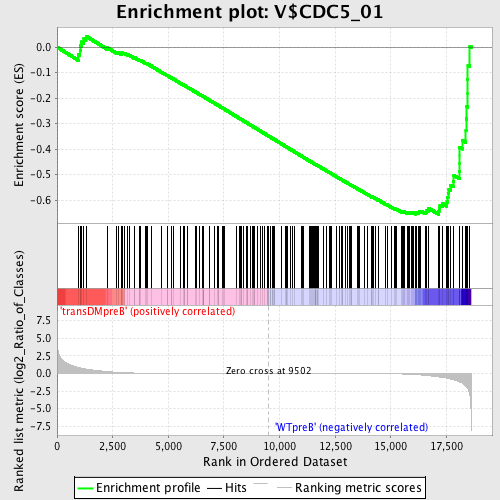
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$CDC5_01 |
| Enrichment Score (ES) | -0.6563666 |
| Normalized Enrichment Score (NES) | -1.6039796 |
| Nominal p-value | 0.0022075055 |
| FDR q-value | 0.07704446 |
| FWER p-Value | 0.255 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SUPT16H | 8541 | 954 | 0.844 | -0.0301 | No | ||
| 2 | ELP3 | 21782 | 1028 | 0.786 | -0.0140 | No | ||
| 3 | PIK3R3 | 5248 | 1045 | 0.772 | 0.0049 | No | ||
| 4 | HMGA1 | 23323 | 1108 | 0.730 | 0.0203 | No | ||
| 5 | PRPS1 | 24233 | 1205 | 0.671 | 0.0323 | No | ||
| 6 | RPA2 | 2330 16057 | 1299 | 0.622 | 0.0432 | No | ||
| 7 | HESX1 | 22070 | 2270 | 0.261 | -0.0028 | No | ||
| 8 | RBMS1 | 14580 | 2663 | 0.162 | -0.0199 | No | ||
| 9 | FNBP1 | 4732 | 2776 | 0.143 | -0.0223 | No | ||
| 10 | TRPS1 | 8195 | 2903 | 0.121 | -0.0260 | No | ||
| 11 | TITF1 | 10180 21062 | 2912 | 0.120 | -0.0234 | No | ||
| 12 | INPP4B | 18557 | 2953 | 0.113 | -0.0227 | No | ||
| 13 | SLC12A8 | 5059 | 3021 | 0.104 | -0.0236 | No | ||
| 14 | PLCB3 | 23799 | 3181 | 0.085 | -0.0301 | No | ||
| 15 | FOXP2 | 4312 17523 8523 | 3267 | 0.076 | -0.0327 | No | ||
| 16 | SLC6A13 | 17300 | 3458 | 0.062 | -0.0415 | No | ||
| 17 | HOXB7 | 666 9110 | 3467 | 0.061 | -0.0403 | No | ||
| 18 | GLI3 | 21703 | 3682 | 0.050 | -0.0506 | No | ||
| 19 | CLDN1 | 22623 | 3712 | 0.049 | -0.0510 | No | ||
| 20 | BCL11B | 7190 | 3768 | 0.046 | -0.0528 | No | ||
| 21 | DUSP14 | 12124 | 3981 | 0.039 | -0.0633 | No | ||
| 22 | SALL1 | 18796 12207 | 4028 | 0.037 | -0.0648 | No | ||
| 23 | ACSL4 | 24043 11936 24042 2614 6940 | 4077 | 0.035 | -0.0665 | No | ||
| 24 | SDC1 | 5560 9953 | 4222 | 0.031 | -0.0735 | No | ||
| 25 | ZFPM2 | 22483 | 4704 | 0.022 | -0.0990 | No | ||
| 26 | GPR87 | 15333 | 4961 | 0.019 | -0.1124 | No | ||
| 27 | HOXD9 | 14982 | 4971 | 0.019 | -0.1124 | No | ||
| 28 | FES | 17779 3949 | 5146 | 0.017 | -0.1214 | No | ||
| 29 | ATRX | 10350 5922 2628 | 5156 | 0.017 | -0.1215 | No | ||
| 30 | ABLIM1 | 3717 3766 3679 | 5229 | 0.016 | -0.1250 | No | ||
| 31 | PROK2 | 17057 | 5549 | 0.014 | -0.1419 | No | ||
| 32 | KCTD15 | 17864 | 5688 | 0.013 | -0.1491 | No | ||
| 33 | MYH6 | 9436 | 5731 | 0.012 | -0.1510 | No | ||
| 34 | CHRNA5 | 8494 | 5741 | 0.012 | -0.1512 | No | ||
| 35 | BIRC4 | 2638 8609 | 5845 | 0.012 | -0.1565 | No | ||
| 36 | DNAJC5B | 15627 | 5851 | 0.012 | -0.1565 | No | ||
| 37 | EMILIN1 | 16891 | 6238 | 0.010 | -0.1772 | No | ||
| 38 | CART1 | 19634 | 6253 | 0.010 | -0.1777 | No | ||
| 39 | BCOR | 12934 2561 | 6416 | 0.009 | -0.1862 | No | ||
| 40 | CITED2 | 5118 14477 | 6512 | 0.009 | -0.1912 | No | ||
| 41 | PCDH8 | 21739 3017 | 6581 | 0.008 | -0.1947 | No | ||
| 42 | ATOH1 | 4419 | 6594 | 0.008 | -0.1951 | No | ||
| 43 | LIN7A | 8424 4230 | 6599 | 0.008 | -0.1951 | No | ||
| 44 | OTP | 21582 | 6828 | 0.007 | -0.2073 | No | ||
| 45 | PRRX1 | 9595 5271 | 6850 | 0.007 | -0.2082 | No | ||
| 46 | RRH | 15168 | 7055 | 0.007 | -0.2191 | No | ||
| 47 | MOS | 16254 | 7229 | 0.006 | -0.2284 | No | ||
| 48 | HOXD10 | 14983 | 7260 | 0.006 | -0.2298 | No | ||
| 49 | PHOX2B | 16524 | 7272 | 0.006 | -0.2303 | No | ||
| 50 | POT1 | 17201 | 7423 | 0.005 | -0.2383 | No | ||
| 51 | TMSB4X | 24005 | 7500 | 0.005 | -0.2423 | No | ||
| 52 | RARA | 5358 | 7529 | 0.005 | -0.2437 | No | ||
| 53 | VANGL1 | 6034 | 8081 | 0.004 | -0.2735 | No | ||
| 54 | EYA1 | 4695 4061 | 8207 | 0.003 | -0.2802 | No | ||
| 55 | PPIL5 | 21255 | 8251 | 0.003 | -0.2824 | No | ||
| 56 | TCF21 | 3437 | 8299 | 0.003 | -0.2849 | No | ||
| 57 | HOXC12 | 22343 | 8309 | 0.003 | -0.2853 | No | ||
| 58 | MYL1 | 4015 | 8370 | 0.003 | -0.2885 | No | ||
| 59 | USP13 | 13047 | 8516 | 0.002 | -0.2963 | No | ||
| 60 | MYL2 | 16713 | 8551 | 0.002 | -0.2981 | No | ||
| 61 | CLASP1 | 14159 | 8552 | 0.002 | -0.2980 | No | ||
| 62 | KLF12 | 4960 2637 21732 9228 | 8710 | 0.002 | -0.3065 | No | ||
| 63 | NAP1L5 | 17134 | 8775 | 0.002 | -0.3099 | No | ||
| 64 | LRFN5 | 6213 | 8843 | 0.002 | -0.3135 | No | ||
| 65 | FGF12 | 1723 22621 | 8850 | 0.002 | -0.3138 | No | ||
| 66 | CALD1 | 4273 8463 | 8892 | 0.001 | -0.3160 | No | ||
| 67 | BTBD3 | 10457 2975 6007 6006 | 9028 | 0.001 | -0.3233 | No | ||
| 68 | RGS1 | 3970 6936 | 9151 | 0.001 | -0.3299 | No | ||
| 69 | POU3F1 | 16093 | 9252 | 0.001 | -0.3353 | No | ||
| 70 | FGF10 | 4719 8962 | 9300 | 0.001 | -0.3378 | No | ||
| 71 | BAI3 | 5580 | 9437 | 0.000 | -0.3452 | No | ||
| 72 | GJA3 | 21807 | 9474 | 0.000 | -0.3471 | No | ||
| 73 | OSR2 | 22492 | 9502 | -0.000 | -0.3486 | No | ||
| 74 | KLHL4 | 24076 | 9506 | -0.000 | -0.3488 | No | ||
| 75 | RORA | 3019 5388 | 9585 | -0.000 | -0.3530 | No | ||
| 76 | ENPP2 | 9548 | 9593 | -0.000 | -0.3533 | No | ||
| 77 | NRXN3 | 2153 5196 | 9658 | -0.000 | -0.3568 | No | ||
| 78 | ODZ1 | 24168 | 9659 | -0.000 | -0.3568 | No | ||
| 79 | SNTG1 | 7717 | 9704 | -0.001 | -0.3592 | No | ||
| 80 | LYN | 16281 | 9749 | -0.001 | -0.3615 | No | ||
| 81 | RCOR1 | 23998 | 9784 | -0.001 | -0.3634 | No | ||
| 82 | IL20 | 13840 | 10096 | -0.002 | -0.3802 | No | ||
| 83 | ROD1 | 6047 2317 | 10271 | -0.002 | -0.3896 | No | ||
| 84 | NRAS | 5191 | 10314 | -0.002 | -0.3918 | No | ||
| 85 | TENC1 | 22349 9927 | 10331 | -0.002 | -0.3926 | No | ||
| 86 | MUSK | 5199 | 10350 | -0.002 | -0.3935 | No | ||
| 87 | MSX1 | 16547 | 10509 | -0.003 | -0.4021 | No | ||
| 88 | TNNI3K | 11495 | 10579 | -0.003 | -0.4057 | No | ||
| 89 | PURA | 9670 | 10677 | -0.003 | -0.4109 | No | ||
| 90 | CDH6 | 22320 | 11000 | -0.004 | -0.4283 | No | ||
| 91 | DGKB | 5722 | 11035 | -0.004 | -0.4300 | No | ||
| 92 | DLL4 | 7040 | 11060 | -0.004 | -0.4312 | No | ||
| 93 | RORB | 10356 | 11322 | -0.005 | -0.4453 | No | ||
| 94 | CHRDL1 | 8180 13504 | 11368 | -0.005 | -0.4476 | No | ||
| 95 | PBX1 | 5224 | 11437 | -0.005 | -0.4511 | No | ||
| 96 | KLF8 | 10885 | 11474 | -0.005 | -0.4530 | No | ||
| 97 | SCN7A | 5413 | 11509 | -0.005 | -0.4547 | No | ||
| 98 | TNNI1 | 14116 | 11563 | -0.005 | -0.4574 | No | ||
| 99 | TNFSF10 | 15626 | 11600 | -0.006 | -0.4592 | No | ||
| 100 | NKX2-3 | 5177 23845 | 11619 | -0.006 | -0.4600 | No | ||
| 101 | IL21 | 12241 | 11628 | -0.006 | -0.4603 | No | ||
| 102 | DNASE1L2 | 960 23092 | 11652 | -0.006 | -0.4614 | No | ||
| 103 | PIK3R1 | 3170 | 11690 | -0.006 | -0.4633 | No | ||
| 104 | ADAMTS9 | 17063 | 11715 | -0.006 | -0.4644 | No | ||
| 105 | NEK6 | 15018 | 11746 | -0.006 | -0.4659 | No | ||
| 106 | BAMBI | 23629 | 11962 | -0.007 | -0.4774 | No | ||
| 107 | HOXD12 | 9113 | 12111 | -0.007 | -0.4852 | No | ||
| 108 | GPC3 | 24151 | 12240 | -0.008 | -0.4920 | No | ||
| 109 | ESRRG | 6447 | 12271 | -0.008 | -0.4934 | No | ||
| 110 | NDRG4 | 10626 | 12331 | -0.008 | -0.4964 | No | ||
| 111 | HOXA11 | 17146 | 12539 | -0.009 | -0.5074 | No | ||
| 112 | PLAG1 | 7148 15949 12161 | 12679 | -0.010 | -0.5146 | No | ||
| 113 | KLF4 | 9230 4962 4963 | 12763 | -0.011 | -0.5189 | No | ||
| 114 | LMNA | 4999 15290 1778 9280 | 12841 | -0.011 | -0.5228 | No | ||
| 115 | FOXA1 | 21060 | 12842 | -0.011 | -0.5225 | No | ||
| 116 | PAX2 | 9530 24416 | 12956 | -0.012 | -0.5283 | No | ||
| 117 | AP1S2 | 8423 4228 | 13045 | -0.012 | -0.5328 | No | ||
| 118 | BMPR2 | 14241 4081 | 13160 | -0.013 | -0.5386 | No | ||
| 119 | ODF1 | 22488 | 13199 | -0.013 | -0.5404 | No | ||
| 120 | CREB5 | 10551 | 13229 | -0.013 | -0.5416 | No | ||
| 121 | HOXC5 | 22340 | 13242 | -0.014 | -0.5419 | No | ||
| 122 | VDR | 5852 10285 | 13499 | -0.016 | -0.5554 | No | ||
| 123 | HIPK1 | 4851 | 13537 | -0.016 | -0.5570 | No | ||
| 124 | PDGFA | 5237 | 13605 | -0.017 | -0.5602 | No | ||
| 125 | NOL4 | 2028 11432 | 13819 | -0.019 | -0.5712 | No | ||
| 126 | COL16A1 | 16066 | 13966 | -0.021 | -0.5786 | No | ||
| 127 | KCNK12 | 9978 | 14138 | -0.024 | -0.5873 | No | ||
| 128 | TLE1 | 5765 10185 15859 | 14152 | -0.024 | -0.5874 | No | ||
| 129 | MGLL | 17368 1145 | 14183 | -0.025 | -0.5884 | No | ||
| 130 | SYT4 | 5566 | 14234 | -0.025 | -0.5904 | No | ||
| 131 | ANGPT1 | 8588 2260 22303 | 14316 | -0.027 | -0.5941 | No | ||
| 132 | SHH | 16581 | 14429 | -0.029 | -0.5995 | No | ||
| 133 | HOXA9 | 17147 9109 1024 | 14436 | -0.029 | -0.5990 | No | ||
| 134 | HOXC6 | 22339 2302 | 14775 | -0.039 | -0.6164 | No | ||
| 135 | NPTX1 | 20126 | 14838 | -0.041 | -0.6187 | No | ||
| 136 | FYN | 3375 3395 20052 | 15046 | -0.049 | -0.6287 | No | ||
| 137 | CUTL1 | 3507 3480 8820 3609 4577 | 15180 | -0.055 | -0.6345 | No | ||
| 138 | ITPKB | 3963 14028 | 15213 | -0.057 | -0.6348 | No | ||
| 139 | UCHL1 | 16834 | 15263 | -0.060 | -0.6359 | No | ||
| 140 | HIVEP3 | 4973 | 15480 | -0.076 | -0.6456 | No | ||
| 141 | ACTR10 | 12132 2047 | 15545 | -0.081 | -0.6470 | No | ||
| 142 | CPNE1 | 11186 | 15562 | -0.083 | -0.6458 | No | ||
| 143 | PRDM13 | 15933 | 15593 | -0.085 | -0.6452 | No | ||
| 144 | FUT11 | 13084 | 15733 | -0.099 | -0.6502 | No | ||
| 145 | SESN3 | 19561 | 15793 | -0.106 | -0.6507 | No | ||
| 146 | SLC27A4 | 15060 | 15820 | -0.109 | -0.6493 | No | ||
| 147 | TGFB3 | 10161 | 15910 | -0.122 | -0.6510 | Yes | ||
| 148 | EGR2 | 8886 | 15957 | -0.132 | -0.6501 | Yes | ||
| 149 | PPARGC1A | 16533 | 16024 | -0.143 | -0.6500 | Yes | ||
| 150 | GPR85 | 7227 1048 | 16111 | -0.166 | -0.6504 | Yes | ||
| 151 | STAG2 | 5521 | 16171 | -0.178 | -0.6491 | Yes | ||
| 152 | YWHAG | 16339 | 16238 | -0.194 | -0.6477 | Yes | ||
| 153 | NDNL2 | 17804 | 16273 | -0.200 | -0.6444 | Yes | ||
| 154 | NLK | 5179 5178 | 16346 | -0.216 | -0.6428 | Yes | ||
| 155 | H3F3B | 4839 | 16546 | -0.270 | -0.6467 | Yes | ||
| 156 | BACH2 | 8648 | 16595 | -0.285 | -0.6420 | Yes | ||
| 157 | HNRPA0 | 13382 | 16677 | -0.308 | -0.6385 | Yes | ||
| 158 | ING3 | 17514 1019 | 16687 | -0.312 | -0.6310 | Yes | ||
| 159 | PRKAG1 | 22135 | 17156 | -0.489 | -0.6438 | Yes | ||
| 160 | PRKACB | 15140 | 17168 | -0.494 | -0.6318 | Yes | ||
| 161 | AICDA | 17295 1087 | 17207 | -0.510 | -0.6208 | Yes | ||
| 162 | CHD2 | 10847 | 17320 | -0.556 | -0.6126 | Yes | ||
| 163 | ARPC5L | 15017 | 17523 | -0.667 | -0.6065 | Yes | ||
| 164 | PBX2 | 1614 23279 | 17554 | -0.692 | -0.5904 | Yes | ||
| 165 | MXI1 | 23813 5139 | 17574 | -0.700 | -0.5735 | Yes | ||
| 166 | B3GNT6 | 23776 | 17610 | -0.729 | -0.5567 | Yes | ||
| 167 | MBNL1 | 1921 15582 | 17687 | -0.781 | -0.5408 | Yes | ||
| 168 | FOXP1 | 4242 | 17816 | -0.882 | -0.5252 | Yes | ||
| 169 | CDKN2C | 15806 2321 | 17833 | -0.892 | -0.5032 | Yes | ||
| 170 | CSNK1G3 | 12873 | 18086 | -1.197 | -0.4862 | Yes | ||
| 171 | SEC63 | 8943 4708 | 18094 | -1.206 | -0.4557 | Yes | ||
| 172 | ARRDC3 | 4191 | 18097 | -1.212 | -0.4247 | Yes | ||
| 173 | MAML1 | 20477 | 18098 | -1.213 | -0.3937 | Yes | ||
| 174 | STAT3 | 5525 9906 | 18210 | -1.378 | -0.3644 | Yes | ||
| 175 | ARNTL | 18119 | 18377 | -1.808 | -0.3271 | Yes | ||
| 176 | CDC42EP3 | 6436 | 18383 | -1.819 | -0.2807 | Yes | ||
| 177 | GFRA1 | 9015 | 18417 | -1.911 | -0.2336 | Yes | ||
| 178 | TMPRSS3 | 23036 | 18456 | -2.125 | -0.1812 | Yes | ||
| 179 | SAMSN1 | 7485 | 18459 | -2.140 | -0.1265 | Yes | ||
| 180 | HSPA2 | 21237 | 18467 | -2.206 | -0.0704 | Yes | ||
| 181 | BCL6 | 22624 | 18554 | -3.060 | 0.0034 | Yes |