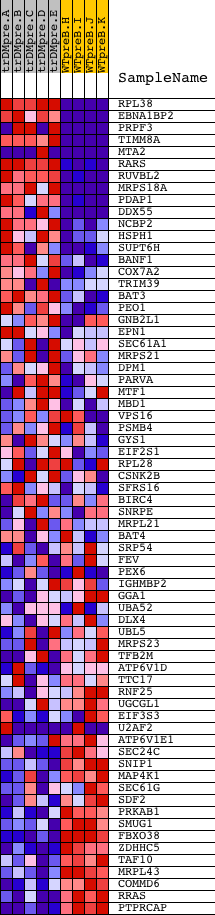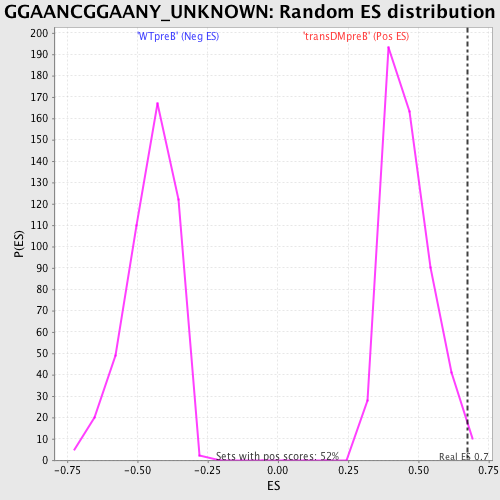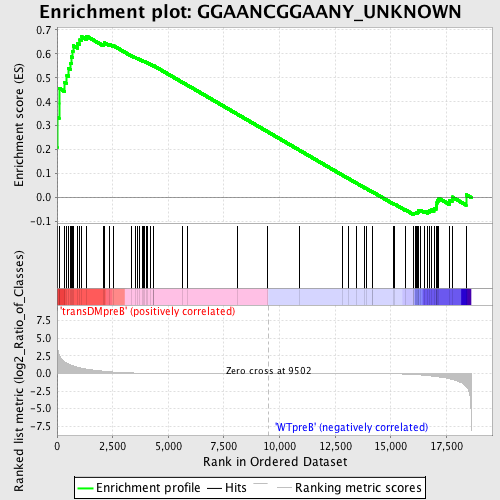
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | GGAANCGGAANY_UNKNOWN |
| Enrichment Score (ES) | 0.67344207 |
| Normalized Enrichment Score (NES) | 1.462038 |
| Nominal p-value | 0.011428571 |
| FDR q-value | 0.3838613 |
| FWER p-Value | 0.957 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL38 | 12562 20606 7475 | 0 | 8.861 | 0.2111 | Yes | ||
| 2 | EBNA1BP2 | 8112 7599 12770 | 6 | 5.130 | 0.3330 | Yes | ||
| 3 | PRPF3 | 12899 12898 1877 7703 | 89 | 2.679 | 0.3924 | Yes | ||
| 4 | TIMM8A | 24062 | 90 | 2.674 | 0.4561 | Yes | ||
| 5 | MTA2 | 23934 18250 | 334 | 1.600 | 0.4811 | Yes | ||
| 6 | RARS | 20496 | 439 | 1.460 | 0.5102 | Yes | ||
| 7 | RUVBL2 | 9766 | 517 | 1.325 | 0.5377 | Yes | ||
| 8 | MRPS18A | 23222 | 609 | 1.182 | 0.5609 | Yes | ||
| 9 | PDAP1 | 16294 | 636 | 1.153 | 0.5870 | Yes | ||
| 10 | DDX55 | 7490 | 690 | 1.090 | 0.6101 | Yes | ||
| 11 | NCBP2 | 12643 | 720 | 1.059 | 0.6338 | Yes | ||
| 12 | HSPH1 | 16282 | 913 | 0.884 | 0.6445 | Yes | ||
| 13 | SUPT6H | 9940 | 1023 | 0.790 | 0.6574 | Yes | ||
| 14 | BANF1 | 6216 10706 3760 | 1090 | 0.740 | 0.6715 | Yes | ||
| 15 | COX7A2 | 19053 | 1324 | 0.610 | 0.6734 | Yes | ||
| 16 | TRIM39 | 22994 | 2080 | 0.319 | 0.6403 | No | ||
| 17 | BAT3 | 5896 | 2111 | 0.309 | 0.6461 | No | ||
| 18 | PEO1 | 5932 | 2342 | 0.242 | 0.6394 | No | ||
| 19 | GNB2L1 | 20911 | 2547 | 0.186 | 0.6329 | No | ||
| 20 | EPN1 | 18401 | 3365 | 0.069 | 0.5905 | No | ||
| 21 | SEC61A1 | 60 | 3505 | 0.059 | 0.5844 | No | ||
| 22 | MRPS21 | 12323 | 3601 | 0.054 | 0.5805 | No | ||
| 23 | DPM1 | 4636 2882 8860 | 3711 | 0.049 | 0.5758 | No | ||
| 24 | PARVA | 18122 | 3832 | 0.044 | 0.5704 | No | ||
| 25 | MTF1 | 5128 9421 | 3883 | 0.042 | 0.5687 | No | ||
| 26 | MBD1 | 1959 23518 1998 2020 | 3906 | 0.041 | 0.5685 | No | ||
| 27 | VPS16 | 2927 14845 2825 | 4005 | 0.038 | 0.5641 | No | ||
| 28 | PSMB4 | 15252 | 4055 | 0.036 | 0.5623 | No | ||
| 29 | GYS1 | 4818 | 4186 | 0.032 | 0.5561 | No | ||
| 30 | EIF2S1 | 4658 | 4327 | 0.029 | 0.5493 | No | ||
| 31 | RPL28 | 5392 | 4339 | 0.029 | 0.5494 | No | ||
| 32 | CSNK2B | 23008 | 4351 | 0.029 | 0.5494 | No | ||
| 33 | SFRS16 | 17943 3840 | 5657 | 0.013 | 0.4794 | No | ||
| 34 | BIRC4 | 2638 8609 | 5845 | 0.012 | 0.4696 | No | ||
| 35 | SNRPE | 9843 | 8094 | 0.004 | 0.3485 | No | ||
| 36 | MRPL21 | 23755 | 9440 | 0.000 | 0.2760 | No | ||
| 37 | BAT4 | 8172 8173 | 10906 | -0.004 | 0.1971 | No | ||
| 38 | SRP54 | 6282 | 12824 | -0.011 | 0.0940 | No | ||
| 39 | FEV | 11153 6434 | 13090 | -0.012 | 0.0800 | No | ||
| 40 | PEX6 | 5900 10331 | 13461 | -0.015 | 0.0604 | No | ||
| 41 | IGHMBP2 | 23945 | 13837 | -0.019 | 0.0407 | No | ||
| 42 | GGA1 | 22432 4197 | 13886 | -0.020 | 0.0386 | No | ||
| 43 | UBA52 | 10239 | 14193 | -0.025 | 0.0226 | No | ||
| 44 | DLX4 | 20282 1225 | 14196 | -0.025 | 0.0231 | No | ||
| 45 | UBL5 | 12299 | 15110 | -0.051 | -0.0249 | No | ||
| 46 | MRPS23 | 12260 | 15172 | -0.055 | -0.0269 | No | ||
| 47 | TFB2M | 4024 9092 4083 | 15652 | -0.090 | -0.0505 | No | ||
| 48 | ATP6V1D | 7872 | 16014 | -0.142 | -0.0666 | No | ||
| 49 | TTC17 | 13219 | 16035 | -0.145 | -0.0642 | No | ||
| 50 | RNF25 | 13922 3929 | 16105 | -0.164 | -0.0640 | No | ||
| 51 | UGCGL1 | 6721 | 16136 | -0.170 | -0.0616 | No | ||
| 52 | EIF3S3 | 12652 | 16200 | -0.185 | -0.0606 | No | ||
| 53 | U2AF2 | 5813 5812 5814 | 16244 | -0.194 | -0.0583 | No | ||
| 54 | ATP6V1E1 | 4423 | 16246 | -0.194 | -0.0537 | No | ||
| 55 | SEC24C | 22086 | 16319 | -0.210 | -0.0526 | No | ||
| 56 | SNIP1 | 16085 | 16504 | -0.259 | -0.0563 | No | ||
| 57 | MAP4K1 | 18313 | 16669 | -0.306 | -0.0579 | No | ||
| 58 | SEC61G | 9795 | 16722 | -0.325 | -0.0530 | No | ||
| 59 | SDF2 | 20762 | 16842 | -0.370 | -0.0506 | No | ||
| 60 | PRKAB1 | 16408 | 16941 | -0.408 | -0.0462 | No | ||
| 61 | SMUG1 | 22109 | 17033 | -0.443 | -0.0405 | No | ||
| 62 | FBXO38 | 23421 | 17048 | -0.450 | -0.0306 | No | ||
| 63 | ZDHHC5 | 14547 | 17071 | -0.460 | -0.0208 | No | ||
| 64 | TAF10 | 10785 | 17099 | -0.469 | -0.0111 | No | ||
| 65 | MRPL43 | 3703 23661 | 17160 | -0.491 | -0.0026 | No | ||
| 66 | COMMD6 | 476 | 17640 | -0.746 | -0.0107 | No | ||
| 67 | RRAS | 18256 | 17755 | -0.834 | 0.0030 | No | ||
| 68 | PTPRCAP | 23767 | 18384 | -1.819 | 0.0125 | No |