Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
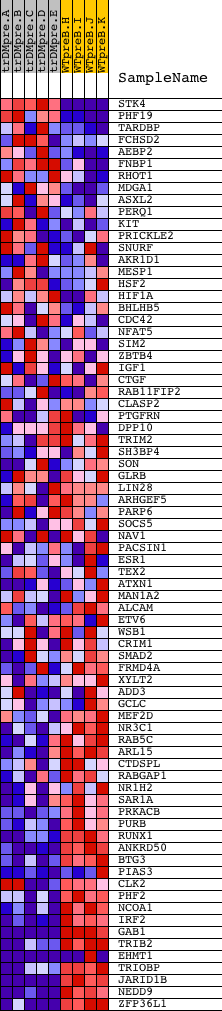
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
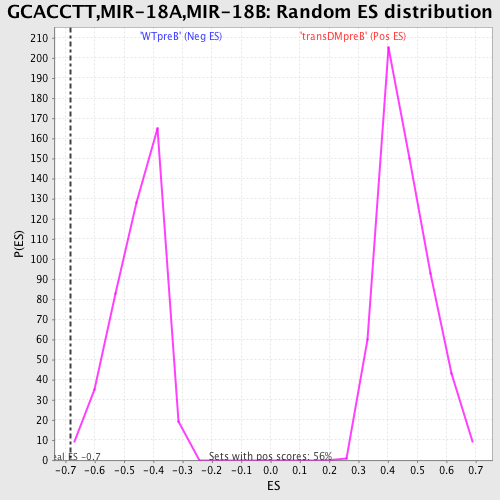
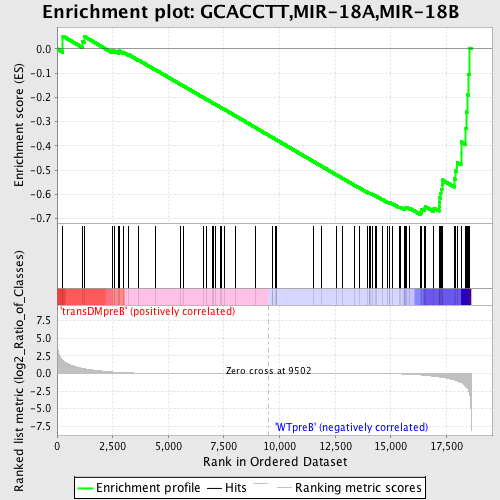
| GeneSet | GCACCTT,MIR-18A,MIR-18B |
| Enrichment Score (ES) | -0.68342674 |
| Normalized Enrichment Score (NES) | -1.5118599 |
| Nominal p-value | 0.0022779044 |
| FDR q-value | 0.14551435 |
| FWER p-Value | 0.742 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | STK4 | 14743 | 262 | 1.794 | 0.0525 | No | ||
| 2 | PHF19 | 13150 | 1144 | 0.706 | 0.0312 | No | ||
| 3 | TARDBP | 10520 6066 | 1241 | 0.651 | 0.0502 | No | ||
| 4 | FCHSD2 | 18172 | 2468 | 0.205 | -0.0083 | No | ||
| 5 | AEBP2 | 4359 | 2598 | 0.173 | -0.0088 | No | ||
| 6 | FNBP1 | 4732 | 2776 | 0.143 | -0.0131 | No | ||
| 7 | RHOT1 | 12227 | 2784 | 0.141 | -0.0082 | No | ||
| 8 | MDGA1 | 13231 | 2974 | 0.110 | -0.0143 | No | ||
| 9 | ASXL2 | 7957 | 3226 | 0.080 | -0.0249 | No | ||
| 10 | PERQ1 | 7168 | 3673 | 0.050 | -0.0471 | No | ||
| 11 | KIT | 16823 | 4403 | 0.027 | -0.0854 | No | ||
| 12 | PRICKLE2 | 10837 | 5566 | 0.013 | -0.1475 | No | ||
| 13 | SNURF | 2251 | 5678 | 0.013 | -0.1530 | No | ||
| 14 | AKR1D1 | 17483 | 6571 | 0.008 | -0.2008 | No | ||
| 15 | MESP1 | 17786 | 6701 | 0.008 | -0.2075 | No | ||
| 16 | HSF2 | 9128 | 7001 | 0.007 | -0.2234 | No | ||
| 17 | HIF1A | 4850 | 7024 | 0.007 | -0.2243 | No | ||
| 18 | BHLHB5 | 15629 | 7107 | 0.006 | -0.2285 | No | ||
| 19 | CDC42 | 4503 8722 4504 2465 | 7364 | 0.005 | -0.2421 | No | ||
| 20 | NFAT5 | 3921 7037 12036 | 7402 | 0.005 | -0.2439 | No | ||
| 21 | SIM2 | 22693 | 7521 | 0.005 | -0.2501 | No | ||
| 22 | ZBTB4 | 13267 | 8020 | 0.004 | -0.2768 | No | ||
| 23 | IGF1 | 3352 9156 3409 | 8900 | 0.001 | -0.3242 | No | ||
| 24 | CTGF | 20068 | 9676 | -0.000 | -0.3659 | No | ||
| 25 | RAB11FIP2 | 23635 | 9695 | -0.001 | -0.3669 | No | ||
| 26 | CLASP2 | 13338 | 9834 | -0.001 | -0.3743 | No | ||
| 27 | PTGFRN | 15224 | 9842 | -0.001 | -0.3747 | No | ||
| 28 | DPP10 | 13850 | 9880 | -0.001 | -0.3766 | No | ||
| 29 | TRIM2 | 13485 8157 | 11527 | -0.005 | -0.4652 | No | ||
| 30 | SH3BP4 | 8270 | 11885 | -0.007 | -0.4842 | No | ||
| 31 | SON | 5473 1657 1684 | 12555 | -0.010 | -0.5199 | No | ||
| 32 | GLRB | 15312 1846 | 12836 | -0.011 | -0.5346 | No | ||
| 33 | LIN28 | 15723 | 13363 | -0.015 | -0.5625 | No | ||
| 34 | ARHGEF5 | 12025 | 13599 | -0.017 | -0.5745 | No | ||
| 35 | PARP6 | 19419 2979 3156 | 13971 | -0.021 | -0.5937 | No | ||
| 36 | SOCS5 | 1619 23141 | 14031 | -0.022 | -0.5961 | No | ||
| 37 | NAV1 | 3978 5681 | 14057 | -0.022 | -0.5966 | No | ||
| 38 | PACSIN1 | 6257 23322 10744 | 14079 | -0.023 | -0.5969 | No | ||
| 39 | ESR1 | 20097 4685 | 14182 | -0.025 | -0.6015 | No | ||
| 40 | TEX2 | 5725 20181 20182 | 14294 | -0.026 | -0.6065 | No | ||
| 41 | ATXN1 | 21477 | 14373 | -0.028 | -0.6097 | No | ||
| 42 | MAN1A2 | 5076 5075 | 14624 | -0.034 | -0.6219 | No | ||
| 43 | ALCAM | 4367 | 14841 | -0.041 | -0.6320 | No | ||
| 44 | ETV6 | 17264 | 14934 | -0.044 | -0.6353 | No | ||
| 45 | WSB1 | 20330 | 14952 | -0.045 | -0.6346 | No | ||
| 46 | CRIM1 | 403 | 15061 | -0.050 | -0.6386 | No | ||
| 47 | SMAD2 | 23511 | 15397 | -0.069 | -0.6541 | No | ||
| 48 | FRMD4A | 5557 | 15426 | -0.071 | -0.6529 | No | ||
| 49 | XYLT2 | 20288 | 15613 | -0.087 | -0.6597 | No | ||
| 50 | ADD3 | 23812 3694 | 15619 | -0.088 | -0.6567 | No | ||
| 51 | GCLC | 19374 | 15672 | -0.093 | -0.6561 | No | ||
| 52 | MEF2D | 9379 | 15716 | -0.097 | -0.6548 | No | ||
| 53 | NR3C1 | 9043 | 15860 | -0.114 | -0.6583 | No | ||
| 54 | RAB5C | 20226 | 16327 | -0.212 | -0.6756 | Yes | ||
| 55 | ARL15 | 21559 | 16367 | -0.222 | -0.6694 | Yes | ||
| 56 | CTDSPL | 19279 | 16390 | -0.227 | -0.6622 | Yes | ||
| 57 | RABGAP1 | 10439 | 16515 | -0.262 | -0.6591 | Yes | ||
| 58 | NR1H2 | 17846 | 16541 | -0.268 | -0.6505 | Yes | ||
| 59 | SAR1A | 20008 | 16939 | -0.407 | -0.6568 | Yes | ||
| 60 | PRKACB | 15140 | 17168 | -0.494 | -0.6507 | Yes | ||
| 61 | PURB | 5335 | 17183 | -0.502 | -0.6328 | Yes | ||
| 62 | RUNX1 | 4481 | 17199 | -0.507 | -0.6148 | Yes | ||
| 63 | ANKRD50 | 533 | 17248 | -0.523 | -0.5980 | Yes | ||
| 64 | BTG3 | 8664 | 17256 | -0.525 | -0.5788 | Yes | ||
| 65 | PIAS3 | 15491 672 1906 | 17302 | -0.547 | -0.5609 | Yes | ||
| 66 | CLK2 | 1883 15544 | 17311 | -0.551 | -0.5409 | Yes | ||
| 67 | PHF2 | 21470 | 17855 | -0.910 | -0.5364 | Yes | ||
| 68 | NCOA1 | 5154 | 17894 | -0.943 | -0.5034 | Yes | ||
| 69 | IRF2 | 18621 | 17974 | -1.043 | -0.4689 | Yes | ||
| 70 | GAB1 | 18828 | 18157 | -1.275 | -0.4314 | Yes | ||
| 71 | TRIB2 | 21102 | 18158 | -1.278 | -0.3839 | Yes | ||
| 72 | EHMT1 | 13401 14670 2756 8090 | 18374 | -1.805 | -0.3285 | Yes | ||
| 73 | TRIOBP | 2280 670 8476 | 18412 | -1.889 | -0.2603 | Yes | ||
| 74 | JARID1B | 14123 | 18428 | -1.963 | -0.1883 | Yes | ||
| 75 | NEDD9 | 3206 21483 | 18492 | -2.359 | -0.1040 | Yes | ||
| 76 | ZFP36L1 | 4453 4454 | 18552 | -2.980 | 0.0035 | Yes |