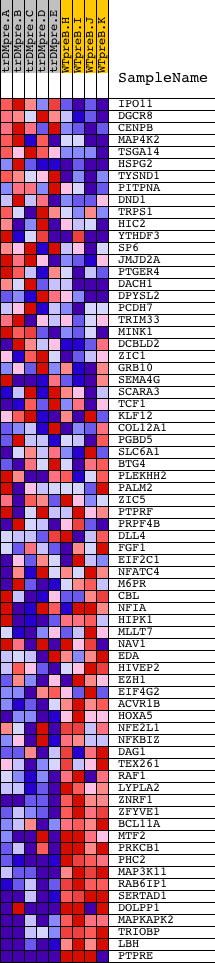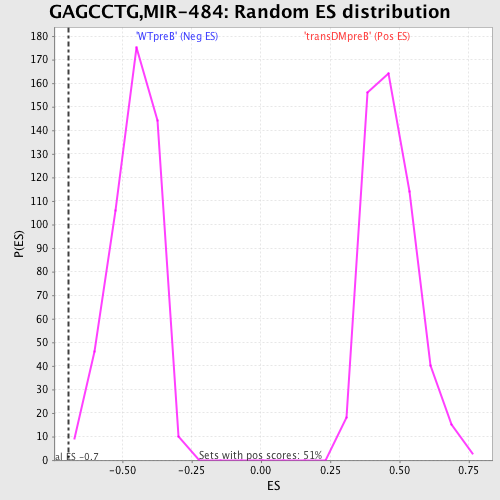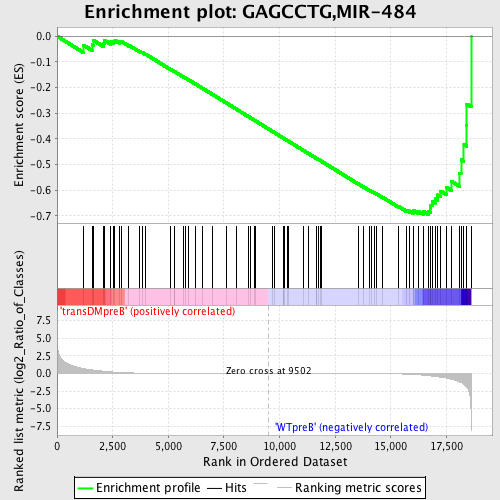
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | GAGCCTG,MIR-484 |
| Enrichment Score (ES) | -0.6946018 |
| Normalized Enrichment Score (NES) | -1.515122 |
| Nominal p-value | 0.006122449 |
| FDR q-value | 0.15782082 |
| FWER p-Value | 0.723 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IPO11 | 21353 | 1198 | 0.675 | -0.0355 | No | ||
| 2 | DGCR8 | 22644 | 1573 | 0.502 | -0.0340 | No | ||
| 3 | CENPB | 14424 | 1618 | 0.483 | -0.0155 | No | ||
| 4 | MAP4K2 | 6457 | 2098 | 0.313 | -0.0278 | No | ||
| 5 | TSGA14 | 17194 | 2125 | 0.305 | -0.0161 | No | ||
| 6 | HSPG2 | 4884 16023 | 2413 | 0.224 | -0.0219 | No | ||
| 7 | TYSND1 | 20007 | 2528 | 0.190 | -0.0198 | No | ||
| 8 | PITPNA | 20778 | 2600 | 0.173 | -0.0162 | No | ||
| 9 | DND1 | 10017 | 2813 | 0.136 | -0.0218 | No | ||
| 10 | TRPS1 | 8195 | 2903 | 0.121 | -0.0213 | No | ||
| 11 | HIC2 | 7185 | 3228 | 0.080 | -0.0354 | No | ||
| 12 | YTHDF3 | 15630 10469 6019 | 3719 | 0.048 | -0.0597 | No | ||
| 13 | SP6 | 8178 13678 13679 | 3826 | 0.044 | -0.0635 | No | ||
| 14 | JMJD2A | 10505 6058 | 3974 | 0.039 | -0.0698 | No | ||
| 15 | PTGER4 | 9649 | 5113 | 0.017 | -0.1304 | No | ||
| 16 | DACH1 | 4595 3371 | 5258 | 0.016 | -0.1375 | No | ||
| 17 | DPYSL2 | 3122 4561 8787 | 5284 | 0.016 | -0.1381 | No | ||
| 18 | PCDH7 | 7029 3460 | 5700 | 0.013 | -0.1600 | No | ||
| 19 | TRIM33 | 8236 13579 15472 8237 | 5789 | 0.012 | -0.1642 | No | ||
| 20 | MINK1 | 20802 | 5922 | 0.011 | -0.1708 | No | ||
| 21 | DCBLD2 | 7855 | 6198 | 0.010 | -0.1852 | No | ||
| 22 | ZIC1 | 19040 | 6535 | 0.008 | -0.2030 | No | ||
| 23 | GRB10 | 4799 | 6979 | 0.007 | -0.2266 | No | ||
| 24 | SEMA4G | 11182 | 7625 | 0.005 | -0.2611 | No | ||
| 25 | SCARA3 | 21780 | 8067 | 0.004 | -0.2848 | No | ||
| 26 | TCF1 | 16416 | 8601 | 0.002 | -0.3134 | No | ||
| 27 | KLF12 | 4960 2637 21732 9228 | 8710 | 0.002 | -0.3192 | No | ||
| 28 | COL12A1 | 3143 19054 | 8891 | 0.001 | -0.3288 | No | ||
| 29 | PGBD5 | 9960 | 8928 | 0.001 | -0.3307 | No | ||
| 30 | SLC6A1 | 10557 | 9678 | -0.000 | -0.3710 | No | ||
| 31 | BTG4 | 19456 | 9767 | -0.001 | -0.3758 | No | ||
| 32 | PLEKHH2 | 23148 | 10163 | -0.002 | -0.3970 | No | ||
| 33 | PALM2 | 10817 2368 | 10166 | -0.002 | -0.3970 | No | ||
| 34 | ZIC5 | 12268 | 10223 | -0.002 | -0.4000 | No | ||
| 35 | PTPRF | 5331 2545 | 10372 | -0.002 | -0.4079 | No | ||
| 36 | PRPF4B | 3208 5295 | 10398 | -0.002 | -0.4091 | No | ||
| 37 | DLL4 | 7040 | 11060 | -0.004 | -0.4446 | No | ||
| 38 | FGF1 | 1994 23447 | 11316 | -0.005 | -0.4581 | No | ||
| 39 | EIF2C1 | 10672 | 11650 | -0.006 | -0.4758 | No | ||
| 40 | NFATC4 | 22002 | 11663 | -0.006 | -0.4762 | No | ||
| 41 | M6PR | 1053 5044 9328 | 11770 | -0.006 | -0.4817 | No | ||
| 42 | CBL | 19154 | 11840 | -0.006 | -0.4851 | No | ||
| 43 | NFIA | 16172 5170 | 11867 | -0.006 | -0.4862 | No | ||
| 44 | HIPK1 | 4851 | 13537 | -0.016 | -0.5755 | No | ||
| 45 | MLLT7 | 24278 | 13782 | -0.019 | -0.5879 | No | ||
| 46 | NAV1 | 3978 5681 | 14057 | -0.022 | -0.6017 | No | ||
| 47 | EDA | 24284 | 14108 | -0.023 | -0.6034 | No | ||
| 48 | HIVEP2 | 9091 | 14249 | -0.026 | -0.6098 | No | ||
| 49 | EZH1 | 20217 | 14344 | -0.027 | -0.6137 | No | ||
| 50 | EIF4G2 | 1908 8892 | 14640 | -0.035 | -0.6281 | No | ||
| 51 | ACVR1B | 4335 22353 | 15353 | -0.065 | -0.6637 | No | ||
| 52 | HOXA5 | 17149 | 15693 | -0.095 | -0.6779 | No | ||
| 53 | NFE2L1 | 9457 | 15839 | -0.111 | -0.6809 | No | ||
| 54 | NFKBIZ | 22575 | 16006 | -0.142 | -0.6838 | Yes | ||
| 55 | DAG1 | 18996 8837 | 16007 | -0.142 | -0.6777 | Yes | ||
| 56 | TEX261 | 17094 5726 | 16259 | -0.197 | -0.6827 | Yes | ||
| 57 | RAF1 | 17035 | 16474 | -0.248 | -0.6835 | Yes | ||
| 58 | LYPLA2 | 15708 | 16680 | -0.309 | -0.6813 | Yes | ||
| 59 | ZNRF1 | 9306 | 16788 | -0.344 | -0.6722 | Yes | ||
| 60 | ZFYVE1 | 10138 | 16800 | -0.350 | -0.6577 | Yes | ||
| 61 | BCL11A | 4691 | 16870 | -0.384 | -0.6449 | Yes | ||
| 62 | MTF2 | 5129 | 16990 | -0.430 | -0.6327 | Yes | ||
| 63 | PRKCB1 | 1693 9574 | 17109 | -0.475 | -0.6186 | Yes | ||
| 64 | PHC2 | 16075 | 17245 | -0.522 | -0.6033 | Yes | ||
| 65 | MAP3K11 | 11163 | 17499 | -0.655 | -0.5887 | Yes | ||
| 66 | RAB6IP1 | 17676 | 17733 | -0.816 | -0.5661 | Yes | ||
| 67 | SERTAD1 | 18326 | 18093 | -1.204 | -0.5335 | Yes | ||
| 68 | DOLPP1 | 2682 15051 | 18156 | -1.273 | -0.4819 | Yes | ||
| 69 | MAPKAPK2 | 13838 | 18287 | -1.556 | -0.4218 | Yes | ||
| 70 | TRIOBP | 2280 670 8476 | 18412 | -1.889 | -0.3470 | Yes | ||
| 71 | LBH | 23160 | 18416 | -1.909 | -0.2648 | Yes | ||
| 72 | PTPRE | 1662 18039 | 18614 | -6.385 | 0.0001 | Yes |