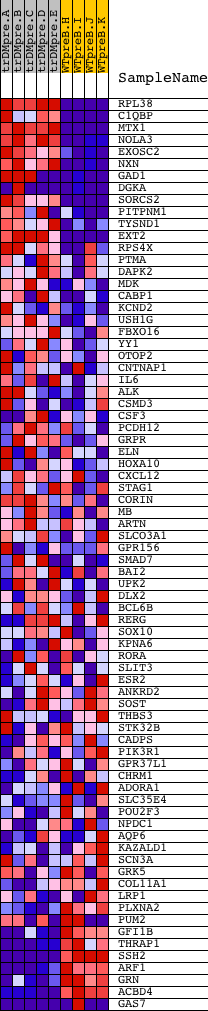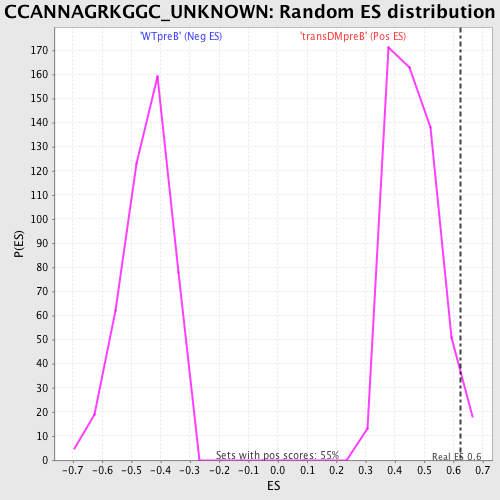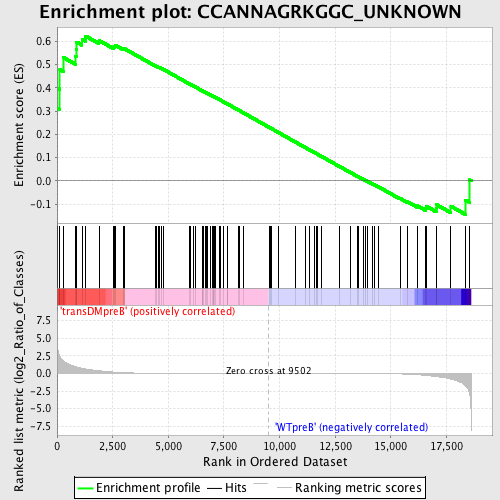
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | CCANNAGRKGGC_UNKNOWN |
| Enrichment Score (ES) | 0.6226834 |
| Normalized Enrichment Score (NES) | 1.3437355 |
| Nominal p-value | 0.039711192 |
| FDR q-value | 0.48741972 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL38 | 12562 20606 7475 | 0 | 8.861 | 0.3119 | Yes | ||
| 2 | C1QBP | 20364 | 99 | 2.537 | 0.3959 | Yes | ||
| 3 | MTX1 | 15282 1918 | 125 | 2.394 | 0.4788 | Yes | ||
| 4 | NOLA3 | 14914 | 280 | 1.717 | 0.5309 | Yes | ||
| 5 | EXOSC2 | 15044 | 836 | 0.949 | 0.5344 | Yes | ||
| 6 | NXN | 5204 | 857 | 0.929 | 0.5660 | Yes | ||
| 7 | GAD1 | 14993 2690 | 876 | 0.913 | 0.5972 | Yes | ||
| 8 | DGKA | 3359 19589 | 1118 | 0.726 | 0.6097 | Yes | ||
| 9 | SORCS2 | 16553 | 1287 | 0.626 | 0.6227 | Yes | ||
| 10 | PITPNM1 | 23770 | 1882 | 0.382 | 0.6041 | No | ||
| 11 | TYSND1 | 20007 | 2528 | 0.190 | 0.5760 | No | ||
| 12 | EXT2 | 14516 | 2596 | 0.174 | 0.5785 | No | ||
| 13 | RPS4X | 9756 | 2634 | 0.168 | 0.5824 | No | ||
| 14 | PTMA | 9657 | 2964 | 0.111 | 0.5686 | No | ||
| 15 | DAPK2 | 19395 | 3037 | 0.101 | 0.5682 | No | ||
| 16 | MDK | 14523 | 4425 | 0.027 | 0.4944 | No | ||
| 17 | CABP1 | 16412 | 4448 | 0.027 | 0.4941 | No | ||
| 18 | KCND2 | 4941 17515 | 4558 | 0.025 | 0.4891 | No | ||
| 19 | USH1G | 20153 | 4597 | 0.024 | 0.4879 | No | ||
| 20 | FBXO16 | 11929 | 4711 | 0.022 | 0.4826 | No | ||
| 21 | YY1 | 10371 | 4772 | 0.022 | 0.4801 | No | ||
| 22 | OTOP2 | 20600 | 5961 | 0.011 | 0.4165 | No | ||
| 23 | CNTNAP1 | 11987 | 6015 | 0.011 | 0.4140 | No | ||
| 24 | IL6 | 16895 | 6131 | 0.010 | 0.4081 | No | ||
| 25 | ALK | 8574 | 6236 | 0.010 | 0.4029 | No | ||
| 26 | CSMD3 | 10735 7938 | 6536 | 0.008 | 0.3870 | No | ||
| 27 | CSF3 | 1394 20671 | 6543 | 0.008 | 0.3870 | No | ||
| 28 | PCDH12 | 7005 | 6584 | 0.008 | 0.3851 | No | ||
| 29 | GRPR | 24015 | 6673 | 0.008 | 0.3807 | No | ||
| 30 | ELN | 8896 | 6704 | 0.008 | 0.3793 | No | ||
| 31 | HOXA10 | 17145 989 | 6759 | 0.008 | 0.3767 | No | ||
| 32 | CXCL12 | 9792 1182 1031 | 6882 | 0.007 | 0.3704 | No | ||
| 33 | STAG1 | 5520 9905 2987 | 7004 | 0.007 | 0.3641 | No | ||
| 34 | CORIN | 16517 | 7031 | 0.007 | 0.3629 | No | ||
| 35 | MB | 22232 | 7046 | 0.007 | 0.3624 | No | ||
| 36 | ARTN | 15783 2430 2532 | 7058 | 0.007 | 0.3620 | No | ||
| 37 | SLCO3A1 | 17796 4237 8430 | 7124 | 0.006 | 0.3587 | No | ||
| 38 | GPR156 | 10753 22763 | 7308 | 0.006 | 0.3491 | No | ||
| 39 | SMAD7 | 23513 | 7342 | 0.006 | 0.3475 | No | ||
| 40 | BAI2 | 2541 2379 16067 | 7497 | 0.005 | 0.3394 | No | ||
| 41 | UPK2 | 19146 | 7637 | 0.005 | 0.3320 | No | ||
| 42 | DLX2 | 14561 353 | 7654 | 0.005 | 0.3313 | No | ||
| 43 | BCL6B | 20373 | 8158 | 0.003 | 0.3043 | No | ||
| 44 | RERG | 16949 | 8198 | 0.003 | 0.3023 | No | ||
| 45 | SOX10 | 22211 | 8392 | 0.003 | 0.2920 | No | ||
| 46 | KPNA6 | 4972 | 9559 | -0.000 | 0.2291 | No | ||
| 47 | RORA | 3019 5388 | 9585 | -0.000 | 0.2278 | No | ||
| 48 | SLIT3 | 20925 | 9626 | -0.000 | 0.2256 | No | ||
| 49 | ESR2 | 21038 | 9948 | -0.001 | 0.2084 | No | ||
| 50 | ANKRD2 | 23853 | 10725 | -0.003 | 0.1666 | No | ||
| 51 | SOST | 20210 | 11179 | -0.004 | 0.1424 | No | ||
| 52 | THBS3 | 15542 1796 | 11344 | -0.005 | 0.1337 | No | ||
| 53 | STK32B | 12255 | 11546 | -0.005 | 0.1230 | No | ||
| 54 | CADPS | 11298 | 11656 | -0.006 | 0.1173 | No | ||
| 55 | PIK3R1 | 3170 | 11690 | -0.006 | 0.1158 | No | ||
| 56 | GPR37L1 | 13825 | 11869 | -0.007 | 0.1064 | No | ||
| 57 | CHRM1 | 23943 | 12673 | -0.010 | 0.0635 | No | ||
| 58 | ADORA1 | 13831 | 13193 | -0.013 | 0.0359 | No | ||
| 59 | SLC35E4 | 20549 | 13509 | -0.016 | 0.0195 | No | ||
| 60 | POU2F3 | 9602 | 13542 | -0.016 | 0.0183 | No | ||
| 61 | NPDC1 | 9479 | 13770 | -0.019 | 0.0067 | No | ||
| 62 | AQP6 | 22364 8621 | 13881 | -0.020 | 0.0015 | No | ||
| 63 | KAZALD1 | 4206 | 13957 | -0.021 | -0.0018 | No | ||
| 64 | SCN3A | 14573 | 14170 | -0.024 | -0.0124 | No | ||
| 65 | GRK5 | 9036 23814 | 14243 | -0.026 | -0.0153 | No | ||
| 66 | COL11A1 | 15442 1937 4540 8762 | 14466 | -0.030 | -0.0263 | No | ||
| 67 | LRP1 | 9284 | 15413 | -0.071 | -0.0748 | No | ||
| 68 | PLXNA2 | 5269 | 15747 | -0.101 | -0.0892 | No | ||
| 69 | PUM2 | 2071 8161 | 16197 | -0.184 | -0.1069 | No | ||
| 70 | GFI1B | 14637 | 16564 | -0.276 | -0.1170 | No | ||
| 71 | THRAP1 | 20309 | 16594 | -0.284 | -0.1085 | No | ||
| 72 | SSH2 | 6202 | 17036 | -0.443 | -0.1167 | No | ||
| 73 | ARF1 | 64 | 17047 | -0.449 | -0.1014 | No | ||
| 74 | GRN | 20640 | 17702 | -0.793 | -0.1088 | No | ||
| 75 | ACBD4 | 20637 | 18357 | -1.760 | -0.0821 | No | ||
| 76 | GAS7 | 20836 1299 | 18533 | -2.729 | 0.0045 | No |