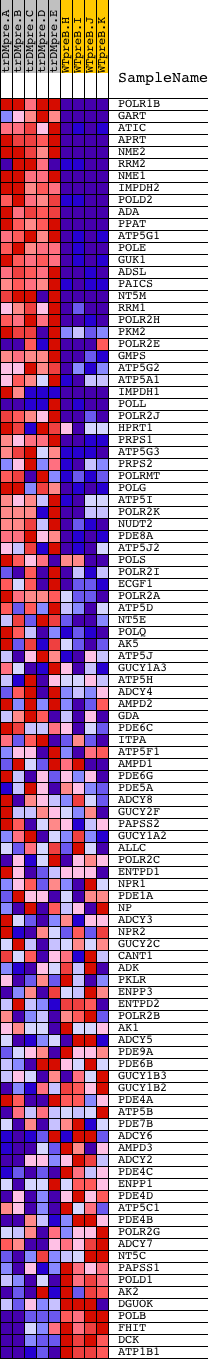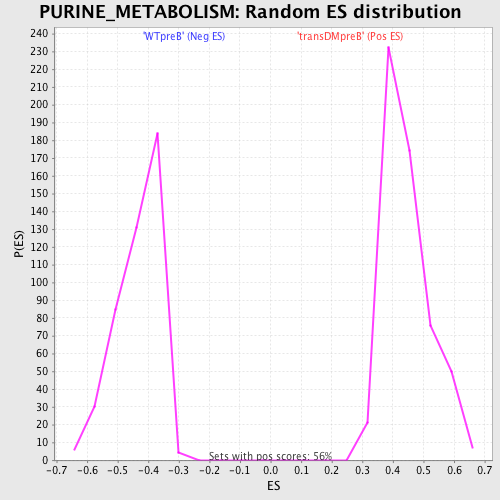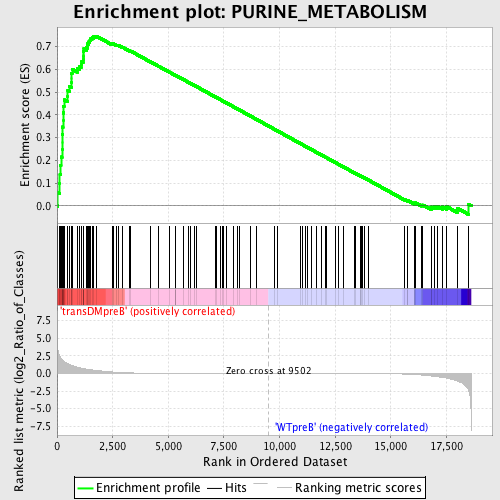
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | PURINE_METABOLISM |
| Enrichment Score (ES) | 0.7458134 |
| Normalized Enrichment Score (NES) | 1.6752264 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025450047 |
| FWER p-Value | 0.032 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POLR1B | 14857 | 32 | 3.400 | 0.0590 | Yes | ||
| 2 | GART | 22543 1754 | 121 | 2.400 | 0.0972 | Yes | ||
| 3 | ATIC | 14231 3968 | 128 | 2.362 | 0.1391 | Yes | ||
| 4 | APRT | 8620 | 144 | 2.254 | 0.1786 | Yes | ||
| 5 | NME2 | 9468 | 175 | 2.112 | 0.2147 | Yes | ||
| 6 | RRM2 | 5401 5400 | 222 | 1.924 | 0.2466 | Yes | ||
| 7 | NME1 | 9467 | 229 | 1.906 | 0.2804 | Yes | ||
| 8 | IMPDH2 | 10730 | 230 | 1.901 | 0.3144 | Yes | ||
| 9 | POLD2 | 20537 | 245 | 1.841 | 0.3465 | Yes | ||
| 10 | ADA | 2703 14361 | 266 | 1.775 | 0.3771 | Yes | ||
| 11 | PPAT | 6081 | 271 | 1.764 | 0.4085 | Yes | ||
| 12 | ATP5G1 | 8636 | 288 | 1.688 | 0.4378 | Yes | ||
| 13 | POLE | 16755 | 314 | 1.622 | 0.4654 | Yes | ||
| 14 | GUK1 | 20432 | 474 | 1.395 | 0.4818 | Yes | ||
| 15 | ADSL | 4358 | 482 | 1.385 | 0.5062 | Yes | ||
| 16 | PAICS | 16820 | 552 | 1.262 | 0.5250 | Yes | ||
| 17 | NT5M | 8345 4175 | 637 | 1.153 | 0.5411 | Yes | ||
| 18 | RRM1 | 18163 | 645 | 1.145 | 0.5611 | Yes | ||
| 19 | POLR2H | 10888 | 646 | 1.144 | 0.5816 | Yes | ||
| 20 | PKM2 | 3642 9573 | 675 | 1.102 | 0.5998 | Yes | ||
| 21 | POLR2E | 3325 19699 | 901 | 0.891 | 0.6036 | Yes | ||
| 22 | GMPS | 15578 | 996 | 0.810 | 0.6130 | Yes | ||
| 23 | ATP5G2 | 12610 | 1079 | 0.751 | 0.6220 | Yes | ||
| 24 | ATP5A1 | 23505 | 1111 | 0.728 | 0.6333 | Yes | ||
| 25 | IMPDH1 | 17197 1131 | 1169 | 0.692 | 0.6426 | Yes | ||
| 26 | POLL | 23658 3688 | 1179 | 0.686 | 0.6544 | Yes | ||
| 27 | POLR2J | 16672 | 1186 | 0.681 | 0.6662 | Yes | ||
| 28 | HPRT1 | 2655 24339 408 | 1187 | 0.681 | 0.6784 | Yes | ||
| 29 | PRPS1 | 24233 | 1205 | 0.671 | 0.6895 | Yes | ||
| 30 | ATP5G3 | 14558 | 1321 | 0.611 | 0.6942 | Yes | ||
| 31 | PRPS2 | 24003 | 1351 | 0.594 | 0.7033 | Yes | ||
| 32 | POLRMT | 19705 | 1380 | 0.578 | 0.7121 | Yes | ||
| 33 | POLG | 17789 | 1419 | 0.562 | 0.7201 | Yes | ||
| 34 | ATP5I | 8637 | 1475 | 0.538 | 0.7267 | Yes | ||
| 35 | POLR2K | 9413 | 1517 | 0.524 | 0.7339 | Yes | ||
| 36 | NUDT2 | 16239 | 1570 | 0.503 | 0.7400 | Yes | ||
| 37 | PDE8A | 18202 | 1648 | 0.471 | 0.7443 | Yes | ||
| 38 | ATP5J2 | 12186 | 1764 | 0.431 | 0.7458 | Yes | ||
| 39 | POLS | 9963 | 2473 | 0.203 | 0.7112 | No | ||
| 40 | POLR2I | 12839 | 2516 | 0.193 | 0.7124 | No | ||
| 41 | ECGF1 | 22160 | 2676 | 0.161 | 0.7067 | No | ||
| 42 | POLR2A | 5394 | 2747 | 0.149 | 0.7055 | No | ||
| 43 | ATP5D | 19949 | 2957 | 0.113 | 0.6963 | No | ||
| 44 | NT5E | 19360 18702 | 3274 | 0.076 | 0.6806 | No | ||
| 45 | POLQ | 13407 22768 | 3312 | 0.073 | 0.6799 | No | ||
| 46 | AK5 | 6042 | 4195 | 0.032 | 0.6328 | No | ||
| 47 | ATP5J | 880 22555 | 4557 | 0.025 | 0.6137 | No | ||
| 48 | GUCY1A3 | 7216 12245 15310 | 5039 | 0.018 | 0.5881 | No | ||
| 49 | ATP5H | 12948 | 5299 | 0.016 | 0.5744 | No | ||
| 50 | ADCY4 | 2980 21818 | 5328 | 0.015 | 0.5731 | No | ||
| 51 | AMPD2 | 1931 15197 | 5330 | 0.015 | 0.5734 | No | ||
| 52 | GDA | 4764 | 5670 | 0.013 | 0.5553 | No | ||
| 53 | PDE6C | 8496 23868 | 5889 | 0.011 | 0.5437 | No | ||
| 54 | ITPA | 9193 | 5993 | 0.011 | 0.5383 | No | ||
| 55 | ATP5F1 | 15212 | 6185 | 0.010 | 0.5282 | No | ||
| 56 | AMPD1 | 5051 | 6248 | 0.010 | 0.5250 | No | ||
| 57 | PDE6G | 20119 | 7120 | 0.006 | 0.4781 | No | ||
| 58 | PDE5A | 15431 | 7151 | 0.006 | 0.4766 | No | ||
| 59 | ADCY8 | 22281 | 7339 | 0.006 | 0.4666 | No | ||
| 60 | GUCY2F | 24045 | 7454 | 0.005 | 0.4605 | No | ||
| 61 | PAPSS2 | 6258 | 7461 | 0.005 | 0.4603 | No | ||
| 62 | GUCY1A2 | 19580 | 7613 | 0.005 | 0.4522 | No | ||
| 63 | ALLC | 21093 2130 | 7948 | 0.004 | 0.4342 | No | ||
| 64 | POLR2C | 9750 | 8110 | 0.003 | 0.4256 | No | ||
| 65 | ENTPD1 | 4495 | 8187 | 0.003 | 0.4215 | No | ||
| 66 | NPR1 | 9480 | 8679 | 0.002 | 0.3951 | No | ||
| 67 | PDE1A | 9541 5234 | 8950 | 0.001 | 0.3805 | No | ||
| 68 | NP | 22027 9597 5273 5274 | 9773 | -0.001 | 0.3361 | No | ||
| 69 | ADCY3 | 21329 894 | 9899 | -0.001 | 0.3294 | No | ||
| 70 | NPR2 | 16230 | 10933 | -0.004 | 0.2736 | No | ||
| 71 | GUCY2C | 1140 16954 | 11013 | -0.004 | 0.2694 | No | ||
| 72 | CANT1 | 13304 | 11171 | -0.004 | 0.2610 | No | ||
| 73 | ADK | 3302 3454 8555 | 11260 | -0.005 | 0.2564 | No | ||
| 74 | PKLR | 1850 15545 | 11427 | -0.005 | 0.2475 | No | ||
| 75 | ENPP3 | 19803 | 11641 | -0.006 | 0.2361 | No | ||
| 76 | ENTPD2 | 8713 | 11872 | -0.007 | 0.2238 | No | ||
| 77 | POLR2B | 16817 | 12059 | -0.007 | 0.2139 | No | ||
| 78 | AK1 | 4363 | 12126 | -0.007 | 0.2104 | No | ||
| 79 | ADCY5 | 10312 | 12491 | -0.009 | 0.1909 | No | ||
| 80 | PDE9A | 23301 | 12652 | -0.010 | 0.1825 | No | ||
| 81 | PDE6B | 16760 | 12880 | -0.011 | 0.1704 | No | ||
| 82 | GUCY1B3 | 15311 | 13349 | -0.014 | 0.1454 | No | ||
| 83 | GUCY1B2 | 21794 | 13402 | -0.015 | 0.1428 | No | ||
| 84 | PDE4A | 3037 19542 3083 3023 | 13615 | -0.017 | 0.1317 | No | ||
| 85 | ATP5B | 19846 | 13623 | -0.017 | 0.1316 | No | ||
| 86 | PDE7B | 19807 | 13687 | -0.018 | 0.1285 | No | ||
| 87 | ADCY6 | 22139 2283 8551 | 13729 | -0.018 | 0.1266 | No | ||
| 88 | AMPD3 | 4383 2110 | 13800 | -0.019 | 0.1232 | No | ||
| 89 | ADCY2 | 21412 | 14014 | -0.022 | 0.1121 | No | ||
| 90 | PDE4C | 8481 | 15624 | -0.088 | 0.0267 | No | ||
| 91 | ENPP1 | 19804 | 15742 | -0.101 | 0.0222 | No | ||
| 92 | PDE4D | 10722 6235 | 15749 | -0.102 | 0.0237 | No | ||
| 93 | ATP5C1 | 8635 | 16066 | -0.152 | 0.0093 | No | ||
| 94 | PDE4B | 9543 | 16079 | -0.158 | 0.0115 | No | ||
| 95 | POLR2G | 23753 | 16110 | -0.165 | 0.0128 | No | ||
| 96 | ADCY7 | 8552 448 | 16369 | -0.222 | 0.0029 | No | ||
| 97 | NT5C | 20151 | 16435 | -0.236 | 0.0036 | No | ||
| 98 | PAPSS1 | 15419 | 16824 | -0.362 | -0.0109 | No | ||
| 99 | POLD1 | 17847 | 16839 | -0.368 | -0.0051 | No | ||
| 100 | AK2 | 8563 2479 16073 | 16960 | -0.419 | -0.0041 | No | ||
| 101 | DGUOK | 17099 1041 | 17097 | -0.468 | -0.0030 | No | ||
| 102 | POLB | 9599 | 17312 | -0.551 | -0.0047 | No | ||
| 103 | FHIT | 21920 | 17514 | -0.663 | -0.0037 | No | ||
| 104 | DCK | 16808 | 17984 | -1.054 | -0.0102 | No | ||
| 105 | ATP1B1 | 4420 | 18501 | -2.479 | 0.0062 | No |