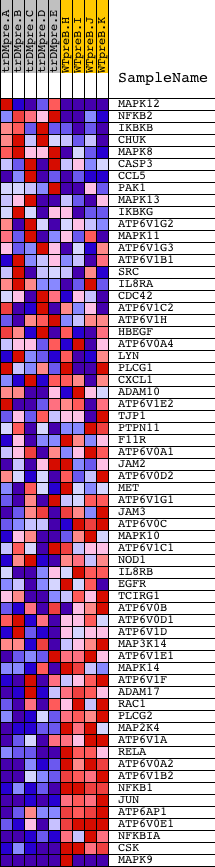
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | HSA05120_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION |
| Enrichment Score (ES) | -0.67274487 |
| Normalized Enrichment Score (NES) | -1.4779408 |
| Nominal p-value | 0.024444444 |
| FDR q-value | 0.29123554 |
| FWER p-Value | 0.971 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAPK12 | 2182 2215 22164 | 1276 | 0.630 | -0.0299 | No | ||
| 2 | NFKB2 | 23810 | 1631 | 0.479 | -0.0195 | No | ||
| 3 | IKBKB | 4907 | 1740 | 0.441 | 0.0019 | No | ||
| 4 | CHUK | 23665 | 1827 | 0.403 | 0.0221 | No | ||
| 5 | MAPK8 | 6459 | 2251 | 0.267 | 0.0157 | No | ||
| 6 | CASP3 | 8693 | 3119 | 0.092 | -0.0254 | No | ||
| 7 | CCL5 | 20320 | 3122 | 0.091 | -0.0199 | No | ||
| 8 | PAK1 | 9527 | 4349 | 0.029 | -0.0842 | No | ||
| 9 | MAPK13 | 23314 | 4628 | 0.024 | -0.0977 | No | ||
| 10 | IKBKG | 2570 2562 4908 | 5311 | 0.016 | -0.1335 | No | ||
| 11 | ATP6V1G2 | 23257 1534 | 5384 | 0.015 | -0.1365 | No | ||
| 12 | MAPK11 | 2264 9618 22163 | 5487 | 0.014 | -0.1411 | No | ||
| 13 | ATP6V1G3 | 14107 | 5824 | 0.012 | -0.1585 | No | ||
| 14 | ATP6V1B1 | 8499 | 6225 | 0.010 | -0.1795 | No | ||
| 15 | SRC | 5507 | 6276 | 0.009 | -0.1816 | No | ||
| 16 | IL8RA | 13925 | 7196 | 0.006 | -0.2307 | No | ||
| 17 | CDC42 | 4503 8722 4504 2465 | 7364 | 0.005 | -0.2394 | No | ||
| 18 | ATP6V1C2 | 21098 | 7828 | 0.004 | -0.2641 | No | ||
| 19 | ATP6V1H | 14301 | 8776 | 0.002 | -0.3150 | No | ||
| 20 | HBEGF | 23457 | 8988 | 0.001 | -0.3263 | No | ||
| 21 | ATP6V0A4 | 8934 | 9568 | -0.000 | -0.3575 | No | ||
| 22 | LYN | 16281 | 9749 | -0.001 | -0.3672 | No | ||
| 23 | PLCG1 | 14753 | 9774 | -0.001 | -0.3684 | No | ||
| 24 | CXCL1 | 9044 | 10817 | -0.003 | -0.4244 | No | ||
| 25 | ADAM10 | 4336 | 10985 | -0.004 | -0.4331 | No | ||
| 26 | ATP6V1E2 | 22880 | 11129 | -0.004 | -0.4406 | No | ||
| 27 | TJP1 | 17803 | 11707 | -0.006 | -0.4713 | No | ||
| 28 | PTPN11 | 5326 16391 9660 | 12084 | -0.007 | -0.4911 | No | ||
| 29 | F11R | 9199 | 12140 | -0.008 | -0.4936 | No | ||
| 30 | ATP6V0A1 | 8640 4424 1197 | 12690 | -0.010 | -0.5226 | No | ||
| 31 | JAM2 | 7428 22718 12519 | 13280 | -0.014 | -0.5535 | No | ||
| 32 | ATP6V0D2 | 15934 | 13572 | -0.016 | -0.5682 | No | ||
| 33 | MET | 17520 | 13603 | -0.017 | -0.5688 | No | ||
| 34 | ATP6V1G1 | 12322 | 13644 | -0.017 | -0.5699 | No | ||
| 35 | JAM3 | 8197 | 13794 | -0.019 | -0.5767 | No | ||
| 36 | ATP6V0C | 8643 | 14389 | -0.028 | -0.6070 | No | ||
| 37 | MAPK10 | 11169 | 14546 | -0.032 | -0.6135 | No | ||
| 38 | ATP6V1C1 | 22487 12329 | 14732 | -0.037 | -0.6211 | No | ||
| 39 | NOD1 | 17141 | 14798 | -0.039 | -0.6222 | No | ||
| 40 | IL8RB | 8752 4533 | 15028 | -0.048 | -0.6316 | No | ||
| 41 | EGFR | 1329 20944 | 15177 | -0.055 | -0.6362 | No | ||
| 42 | TCIRG1 | 3700 3752 23951 | 15305 | -0.062 | -0.6392 | No | ||
| 43 | ATP6V0B | 15786 | 15928 | -0.126 | -0.6650 | Yes | ||
| 44 | ATP6V0D1 | 3895 8639 3920 | 15931 | -0.127 | -0.6572 | Yes | ||
| 45 | ATP6V1D | 7872 | 16014 | -0.142 | -0.6529 | Yes | ||
| 46 | MAP3K14 | 11998 | 16026 | -0.144 | -0.6446 | Yes | ||
| 47 | ATP6V1E1 | 4423 | 16246 | -0.194 | -0.6445 | Yes | ||
| 48 | MAPK14 | 23313 | 16271 | -0.200 | -0.6334 | Yes | ||
| 49 | ATP6V1F | 12291 | 16315 | -0.209 | -0.6229 | Yes | ||
| 50 | ADAM17 | 4343 | 16871 | -0.384 | -0.6291 | Yes | ||
| 51 | RAC1 | 16302 | 17052 | -0.451 | -0.6110 | Yes | ||
| 52 | PLCG2 | 18453 | 17084 | -0.464 | -0.5841 | Yes | ||
| 53 | MAP2K4 | 20405 | 17227 | -0.515 | -0.5600 | Yes | ||
| 54 | ATP6V1A | 8638 | 17552 | -0.690 | -0.5349 | Yes | ||
| 55 | RELA | 23783 | 17583 | -0.705 | -0.4931 | Yes | ||
| 56 | ATP6V0A2 | 5760 | 17586 | -0.708 | -0.4496 | Yes | ||
| 57 | ATP6V1B2 | 18599 | 17669 | -0.769 | -0.4066 | Yes | ||
| 58 | NFKB1 | 15160 | 17682 | -0.778 | -0.3593 | Yes | ||
| 59 | JUN | 15832 | 17701 | -0.791 | -0.3115 | Yes | ||
| 60 | ATP6AP1 | 24296 | 17782 | -0.854 | -0.2632 | Yes | ||
| 61 | ATP6V0E1 | 23326 | 17864 | -0.916 | -0.2111 | Yes | ||
| 62 | NFKBIA | 21065 | 18048 | -1.142 | -0.1506 | Yes | ||
| 63 | CSK | 8805 | 18077 | -1.181 | -0.0793 | Yes | ||
| 64 | MAPK9 | 1233 20903 1383 | 18355 | -1.758 | 0.0141 | Yes |