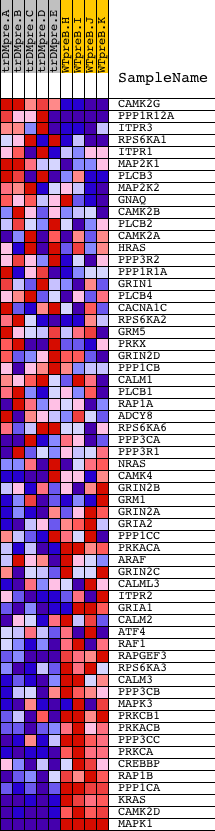
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
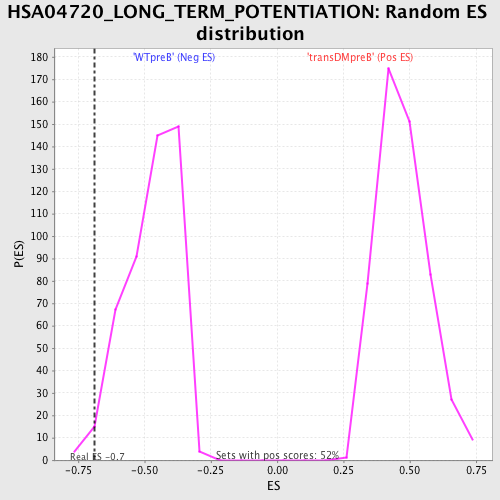
| GeneSet | HSA04720_LONG_TERM_POTENTIATION |
| Enrichment Score (ES) | -0.6893486 |
| Normalized Enrichment Score (NES) | -1.464199 |
| Nominal p-value | 0.021052632 |
| FDR q-value | 0.30317634 |
| FWER p-Value | 0.987 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CAMK2G | 21905 | 1141 | 0.708 | -0.0211 | No | ||
| 2 | PPP1R12A | 3354 19886 | 1508 | 0.525 | -0.0110 | No | ||
| 3 | ITPR3 | 9195 | 1704 | 0.453 | 0.0043 | No | ||
| 4 | RPS6KA1 | 15725 | 2384 | 0.231 | -0.0191 | No | ||
| 5 | ITPR1 | 17341 | 2532 | 0.190 | -0.0162 | No | ||
| 6 | MAP2K1 | 19082 | 2640 | 0.166 | -0.0125 | No | ||
| 7 | PLCB3 | 23799 | 3181 | 0.085 | -0.0368 | No | ||
| 8 | MAP2K2 | 19933 | 3325 | 0.072 | -0.0404 | No | ||
| 9 | GNAQ | 4786 23909 3685 | 3787 | 0.045 | -0.0626 | No | ||
| 10 | CAMK2B | 20536 | 4149 | 0.033 | -0.0802 | No | ||
| 11 | PLCB2 | 5262 | 4304 | 0.030 | -0.0868 | No | ||
| 12 | CAMK2A | 2024 23541 1980 | 4715 | 0.022 | -0.1076 | No | ||
| 13 | HRAS | 4868 | 4722 | 0.022 | -0.1067 | No | ||
| 14 | PPP3R2 | 9612 | 4969 | 0.019 | -0.1188 | No | ||
| 15 | PPP1R1A | 22102 | 5165 | 0.017 | -0.1284 | No | ||
| 16 | GRIN1 | 9041 4804 | 5293 | 0.016 | -0.1343 | No | ||
| 17 | PLCB4 | 9586 | 6139 | 0.010 | -0.1793 | No | ||
| 18 | CACNA1C | 1158 1166 24464 4463 8674 | 6269 | 0.010 | -0.1857 | No | ||
| 19 | RPS6KA2 | 9759 9758 | 6361 | 0.009 | -0.1901 | No | ||
| 20 | GRM5 | 8426 | 6469 | 0.009 | -0.1953 | No | ||
| 21 | PRKX | 9619 5290 | 6505 | 0.009 | -0.1967 | No | ||
| 22 | GRIN2D | 9042 | 6645 | 0.008 | -0.2038 | No | ||
| 23 | PPP1CB | 5282 3529 3504 | 6891 | 0.007 | -0.2166 | No | ||
| 24 | CALM1 | 21184 | 6895 | 0.007 | -0.2163 | No | ||
| 25 | PLCB1 | 14832 2821 | 7050 | 0.007 | -0.2243 | No | ||
| 26 | RAP1A | 8467 | 7144 | 0.006 | -0.2289 | No | ||
| 27 | ADCY8 | 22281 | 7339 | 0.006 | -0.2391 | No | ||
| 28 | RPS6KA6 | 12458 | 7556 | 0.005 | -0.2504 | No | ||
| 29 | PPP3CA | 1863 5284 | 9030 | 0.001 | -0.3297 | No | ||
| 30 | PPP3R1 | 9611 489 | 10273 | -0.002 | -0.3965 | No | ||
| 31 | NRAS | 5191 | 10314 | -0.002 | -0.3986 | No | ||
| 32 | CAMK4 | 4473 | 10887 | -0.004 | -0.4292 | No | ||
| 33 | GRIN2B | 16957 | 11537 | -0.005 | -0.4639 | No | ||
| 34 | GRM1 | 19822 | 12959 | -0.012 | -0.5398 | No | ||
| 35 | GRIN2A | 22668 | 13570 | -0.016 | -0.5717 | No | ||
| 36 | GRIA2 | 4802 | 14049 | -0.022 | -0.5962 | No | ||
| 37 | PPP1CC | 9609 5283 | 14236 | -0.025 | -0.6048 | No | ||
| 38 | PRKACA | 18549 3844 | 14320 | -0.027 | -0.6078 | No | ||
| 39 | ARAF | 24367 | 14795 | -0.039 | -0.6311 | No | ||
| 40 | GRIN2C | 20155 | 15004 | -0.047 | -0.6396 | No | ||
| 41 | CALML3 | 21553 | 15452 | -0.074 | -0.6595 | No | ||
| 42 | ITPR2 | 9194 | 15655 | -0.091 | -0.6652 | No | ||
| 43 | GRIA1 | 20877 4801 9040 | 15689 | -0.094 | -0.6616 | No | ||
| 44 | CALM2 | 8681 | 16194 | -0.184 | -0.6783 | Yes | ||
| 45 | ATF4 | 22417 2239 | 16400 | -0.229 | -0.6763 | Yes | ||
| 46 | RAF1 | 17035 | 16474 | -0.248 | -0.6661 | Yes | ||
| 47 | RAPGEF3 | 22146 | 16578 | -0.277 | -0.6559 | Yes | ||
| 48 | RPS6KA3 | 8490 | 16636 | -0.295 | -0.6421 | Yes | ||
| 49 | CALM3 | 8682 | 16904 | -0.395 | -0.6341 | Yes | ||
| 50 | PPP3CB | 5285 | 16962 | -0.419 | -0.6133 | Yes | ||
| 51 | MAPK3 | 6458 11170 | 17011 | -0.437 | -0.5910 | Yes | ||
| 52 | PRKCB1 | 1693 9574 | 17109 | -0.475 | -0.5692 | Yes | ||
| 53 | PRKACB | 15140 | 17168 | -0.494 | -0.5441 | Yes | ||
| 54 | PPP3CC | 21763 | 17176 | -0.498 | -0.5161 | Yes | ||
| 55 | PRKCA | 20174 | 17237 | -0.520 | -0.4897 | Yes | ||
| 56 | CREBBP | 22682 8783 | 17641 | -0.746 | -0.4689 | Yes | ||
| 57 | RAP1B | 10083 | 17748 | -0.823 | -0.4277 | Yes | ||
| 58 | PPP1CA | 9608 3756 914 909 | 18001 | -1.084 | -0.3796 | Yes | ||
| 59 | KRAS | 9247 | 18298 | -1.598 | -0.3045 | Yes | ||
| 60 | CAMK2D | 4232 | 18399 | -1.851 | -0.2044 | Yes | ||
| 61 | MAPK1 | 1642 11167 | 18579 | -3.793 | 0.0020 | Yes |