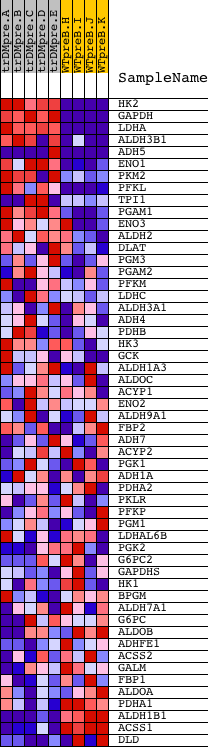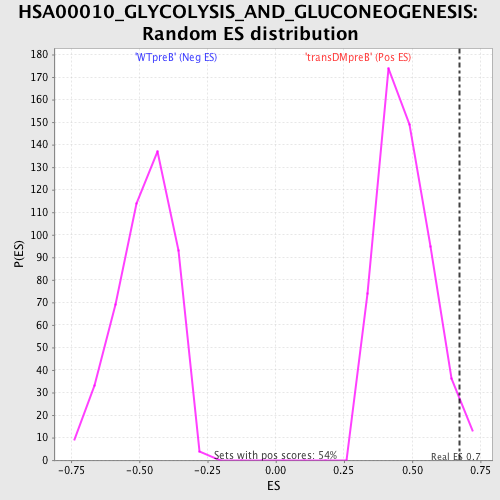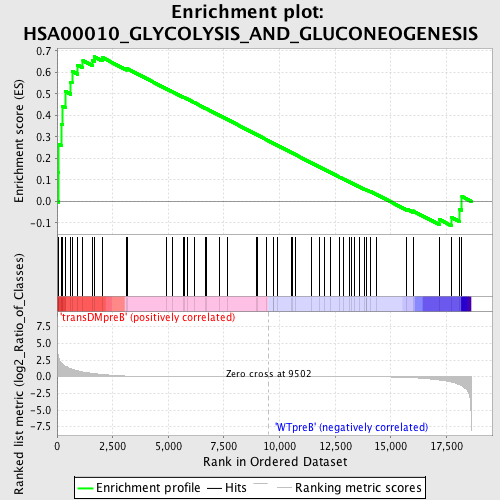
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | HSA00010_GLYCOLYSIS_AND_GLUCONEOGENESIS |
| Enrichment Score (ES) | 0.67196375 |
| Normalized Enrichment Score (NES) | 1.4185599 |
| Nominal p-value | 0.027726432 |
| FDR q-value | 0.5657911 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HK2 | 17106 | 74 | 2.880 | 0.1341 | Yes | ||
| 2 | GAPDH | 348 348 9002 | 83 | 2.748 | 0.2653 | Yes | ||
| 3 | LDHA | 9269 | 190 | 2.057 | 0.3582 | Yes | ||
| 4 | ALDH3B1 | 12569 23949 | 247 | 1.838 | 0.4433 | Yes | ||
| 5 | ADH5 | 8554 15404 | 364 | 1.556 | 0.5116 | Yes | ||
| 6 | ENO1 | 8903 | 611 | 1.181 | 0.5550 | Yes | ||
| 7 | PKM2 | 3642 9573 | 675 | 1.102 | 0.6044 | Yes | ||
| 8 | PFKL | 9553 | 916 | 0.881 | 0.6337 | Yes | ||
| 9 | TPI1 | 5795 10212 | 1160 | 0.698 | 0.6541 | Yes | ||
| 10 | PGAM1 | 9556 | 1578 | 0.500 | 0.6556 | Yes | ||
| 11 | ENO3 | 8905 | 1683 | 0.459 | 0.6720 | Yes | ||
| 12 | ALDH2 | 16384 | 2040 | 0.328 | 0.6685 | No | ||
| 13 | DLAT | 19123 | 3116 | 0.092 | 0.6150 | No | ||
| 14 | PGM3 | 19045 | 3153 | 0.088 | 0.6173 | No | ||
| 15 | PGAM2 | 20539 | 4923 | 0.020 | 0.5229 | No | ||
| 16 | PFKM | 5243 9554 | 5164 | 0.017 | 0.5108 | No | ||
| 17 | LDHC | 18235 2235 | 5685 | 0.013 | 0.4834 | No | ||
| 18 | ALDH3A1 | 20854 | 5707 | 0.012 | 0.4829 | No | ||
| 19 | ADH4 | 15405 | 5840 | 0.012 | 0.4763 | No | ||
| 20 | PDHB | 12670 7548 | 6158 | 0.010 | 0.4597 | No | ||
| 21 | HK3 | 3224 477 | 6649 | 0.008 | 0.4337 | No | ||
| 22 | GCK | 20535 | 6732 | 0.008 | 0.4296 | No | ||
| 23 | ALDH1A3 | 17802 | 7278 | 0.006 | 0.4006 | No | ||
| 24 | ALDOC | 20759 311 | 7658 | 0.005 | 0.3804 | No | ||
| 25 | ACYP1 | 21019 | 8943 | 0.001 | 0.3113 | No | ||
| 26 | ENO2 | 8904 | 8991 | 0.001 | 0.3088 | No | ||
| 27 | ALDH9A1 | 14064 | 9402 | 0.000 | 0.2867 | No | ||
| 28 | FBP2 | 21422 | 9716 | -0.001 | 0.2699 | No | ||
| 29 | ADH7 | 15408 | 9731 | -0.001 | 0.2692 | No | ||
| 30 | ACYP2 | 20508 | 9920 | -0.001 | 0.2591 | No | ||
| 31 | PGK1 | 9557 5244 | 10544 | -0.003 | 0.2257 | No | ||
| 32 | ADH1A | 15406 | 10601 | -0.003 | 0.2228 | No | ||
| 33 | PDHA2 | 15153 | 10692 | -0.003 | 0.2181 | No | ||
| 34 | PKLR | 1850 15545 | 11427 | -0.005 | 0.1788 | No | ||
| 35 | PFKP | 3267 3168 7118 | 11796 | -0.006 | 0.1592 | No | ||
| 36 | PGM1 | 16841 | 12021 | -0.007 | 0.1475 | No | ||
| 37 | LDHAL6B | 8386 | 12270 | -0.008 | 0.1345 | No | ||
| 38 | PGK2 | 15379 | 12707 | -0.010 | 0.1115 | No | ||
| 39 | G6PC2 | 2957 14999 | 12871 | -0.011 | 0.1033 | No | ||
| 40 | GAPDHS | 4752 | 13127 | -0.013 | 0.0902 | No | ||
| 41 | HK1 | 4854 | 13249 | -0.014 | 0.0843 | No | ||
| 42 | BPGM | 17489 | 13387 | -0.015 | 0.0776 | No | ||
| 43 | ALDH7A1 | 23435 | 13579 | -0.016 | 0.0681 | No | ||
| 44 | G6PC | 20656 | 13811 | -0.019 | 0.0566 | No | ||
| 45 | ALDOB | 10494 | 13910 | -0.021 | 0.0523 | No | ||
| 46 | ADHFE1 | 4049 8008 | 13927 | -0.021 | 0.0524 | No | ||
| 47 | ACSS2 | 12242 2684 | 14100 | -0.023 | 0.0443 | No | ||
| 48 | GALM | 23152 | 14364 | -0.028 | 0.0314 | No | ||
| 49 | FBP1 | 21423 | 15719 | -0.097 | -0.0369 | No | ||
| 50 | ALDOA | 8572 | 15996 | -0.139 | -0.0451 | No | ||
| 51 | PDHA1 | 24020 | 17188 | -0.502 | -0.0852 | No | ||
| 52 | ALDH1B1 | 16219 | 17740 | -0.818 | -0.0756 | No | ||
| 53 | ACSS1 | 12729 | 18096 | -1.212 | -0.0367 | No | ||
| 54 | DLD | 2097 21090 | 18196 | -1.348 | 0.0226 | No |