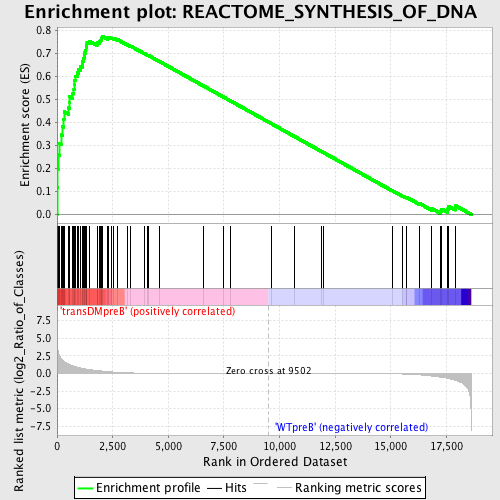
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | REACTOME_SYNTHESIS_OF_DNA |
| Enrichment Score (ES) | 0.77590775 |
| Normalized Enrichment Score (NES) | 1.6278206 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.039365426 |
| FWER p-Value | 0.157 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMC4 | 2141 17915 | 4 | 5.704 | 0.1171 | Yes | ||
| 2 | PSMD8 | 7166 | 21 | 3.807 | 0.1946 | Yes | ||
| 3 | PSMB9 | 23021 | 43 | 3.226 | 0.2598 | Yes | ||
| 4 | PSMD7 | 18752 3825 | 102 | 2.516 | 0.3084 | Yes | ||
| 5 | MCM7 | 9372 3568 | 178 | 2.099 | 0.3475 | Yes | ||
| 6 | POLD2 | 20537 | 245 | 1.841 | 0.3818 | Yes | ||
| 7 | RFC1 | 16527 | 282 | 1.712 | 0.4151 | Yes | ||
| 8 | POLE | 16755 | 314 | 1.622 | 0.4468 | Yes | ||
| 9 | RFC5 | 13005 7791 | 515 | 1.327 | 0.4633 | Yes | ||
| 10 | RFC4 | 1735 22627 | 560 | 1.250 | 0.4867 | Yes | ||
| 11 | PSMD2 | 10137 5724 | 565 | 1.244 | 0.5120 | Yes | ||
| 12 | PSMC5 | 1348 20624 | 708 | 1.076 | 0.5265 | Yes | ||
| 13 | MCM3 | 13991 | 755 | 1.026 | 0.5451 | Yes | ||
| 14 | POLA2 | 23988 | 781 | 1.001 | 0.5644 | Yes | ||
| 15 | PSMA7 | 14318 | 787 | 0.995 | 0.5846 | Yes | ||
| 16 | MCM2 | 17074 | 826 | 0.958 | 0.6022 | Yes | ||
| 17 | PSMD3 | 5803 | 905 | 0.888 | 0.6163 | Yes | ||
| 18 | PSME3 | 20657 | 957 | 0.841 | 0.6309 | Yes | ||
| 19 | RPA1 | 20349 | 1036 | 0.778 | 0.6426 | Yes | ||
| 20 | PRIM1 | 19847 | 1125 | 0.723 | 0.6528 | Yes | ||
| 21 | POLD3 | 17742 | 1142 | 0.708 | 0.6665 | Yes | ||
| 22 | PSMA4 | 11179 | 1176 | 0.687 | 0.6788 | Yes | ||
| 23 | PSMD12 | 20621 | 1217 | 0.663 | 0.6903 | Yes | ||
| 24 | MCM5 | 18564 | 1236 | 0.655 | 0.7028 | Yes | ||
| 25 | PCNA | 9535 | 1278 | 0.629 | 0.7135 | Yes | ||
| 26 | RPA2 | 2330 16057 | 1299 | 0.622 | 0.7253 | Yes | ||
| 27 | FEN1 | 8961 | 1315 | 0.613 | 0.7371 | Yes | ||
| 28 | RPA3 | 12667 | 1341 | 0.601 | 0.7481 | Yes | ||
| 29 | PSME1 | 5300 9637 | 1444 | 0.550 | 0.7539 | Yes | ||
| 30 | CDK2 | 3438 3373 19592 3322 | 1795 | 0.415 | 0.7436 | Yes | ||
| 31 | PSMD14 | 15005 | 1829 | 0.402 | 0.7501 | Yes | ||
| 32 | PSMB5 | 9633 | 1912 | 0.373 | 0.7533 | Yes | ||
| 33 | PSMC2 | 16909 | 1953 | 0.359 | 0.7585 | Yes | ||
| 34 | PSMD10 | 2578 2597 24046 | 1991 | 0.348 | 0.7637 | Yes | ||
| 35 | PSMB6 | 9634 | 2015 | 0.337 | 0.7694 | Yes | ||
| 36 | ORC5L | 11173 3595 | 2023 | 0.336 | 0.7759 | Yes | ||
| 37 | PSMA5 | 6464 | 2278 | 0.259 | 0.7675 | No | ||
| 38 | RFC3 | 12786 | 2294 | 0.254 | 0.7720 | No | ||
| 39 | PSMD9 | 3461 7393 | 2454 | 0.211 | 0.7677 | No | ||
| 40 | PSMC1 | 9635 | 2542 | 0.187 | 0.7669 | No | ||
| 41 | PSMC6 | 7379 | 2726 | 0.153 | 0.7602 | No | ||
| 42 | PSMC3 | 9636 | 3174 | 0.086 | 0.7378 | No | ||
| 43 | PSMB3 | 11180 | 3314 | 0.073 | 0.7319 | No | ||
| 44 | PSMD4 | 15251 | 3907 | 0.041 | 0.7008 | No | ||
| 45 | PSMB4 | 15252 | 4055 | 0.036 | 0.6936 | No | ||
| 46 | PSMA2 | 9631 | 4099 | 0.035 | 0.6920 | No | ||
| 47 | ORC4L | 11172 6460 | 4586 | 0.024 | 0.6663 | No | ||
| 48 | CDC45L | 22642 1752 | 6572 | 0.008 | 0.5595 | No | ||
| 49 | PSMD5 | 2774 14612 | 7473 | 0.005 | 0.5111 | No | ||
| 50 | POLE2 | 21053 | 7815 | 0.004 | 0.4928 | No | ||
| 51 | POLA1 | 24112 | 9653 | -0.000 | 0.3938 | No | ||
| 52 | PSMA1 | 1627 17669 | 10653 | -0.003 | 0.3400 | No | ||
| 53 | PSMA6 | 21270 | 11864 | -0.006 | 0.2749 | No | ||
| 54 | PSMA3 | 9632 5298 | 11968 | -0.007 | 0.2695 | No | ||
| 55 | ORC2L | 385 13949 | 15052 | -0.049 | 0.1043 | No | ||
| 56 | PSMD11 | 12772 7600 | 15541 | -0.081 | 0.0797 | No | ||
| 57 | MCM6 | 4000 13845 4119 | 15695 | -0.095 | 0.0734 | No | ||
| 58 | ORC1L | 327 16144 | 15713 | -0.097 | 0.0745 | No | ||
| 59 | PSMB1 | 23118 | 16286 | -0.203 | 0.0478 | No | ||
| 60 | POLD1 | 17847 | 16839 | -0.368 | 0.0256 | No | ||
| 61 | PSMB10 | 5299 18761 | 17236 | -0.520 | 0.0150 | No | ||
| 62 | LIG1 | 18388 1749 1493 | 17285 | -0.539 | 0.0235 | No | ||
| 63 | MCM4 | 22655 1708 | 17543 | -0.687 | 0.0237 | No | ||
| 64 | RB1 | 21754 | 17584 | -0.706 | 0.0361 | No | ||
| 65 | PSMB2 | 2324 16078 | 17897 | -0.947 | 0.0388 | No |

