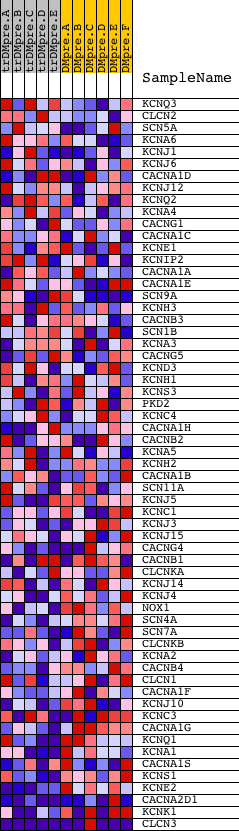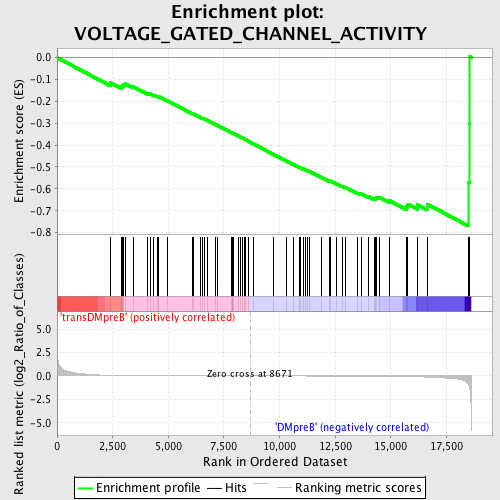
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | VOLTAGE_GATED_CHANNEL_ACTIVITY |
| Enrichment Score (ES) | -0.7697038 |
| Normalized Enrichment Score (NES) | -1.6987226 |
| Nominal p-value | 0.0021413276 |
| FDR q-value | 0.36397782 |
| FWER p-Value | 0.259 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KCNQ3 | 22279 | 2398 | 0.062 | -0.1158 | No | ||
| 2 | CLCN2 | 22637 | 2892 | 0.040 | -0.1337 | No | ||
| 3 | SCN5A | 5412 | 2936 | 0.039 | -0.1277 | No | ||
| 4 | KCNA6 | 664 16990 665 4938 | 2995 | 0.037 | -0.1229 | No | ||
| 5 | KCNJ1 | 3121 12110 | 3090 | 0.034 | -0.1206 | No | ||
| 6 | KCNJ6 | 22533 386 | 3429 | 0.026 | -0.1332 | No | ||
| 7 | CACNA1D | 4464 8675 | 4052 | 0.018 | -0.1630 | No | ||
| 8 | KCNJ12 | 20855 | 4176 | 0.016 | -0.1661 | No | ||
| 9 | KCNQ2 | 164 162 163 | 4329 | 0.015 | -0.1712 | No | ||
| 10 | KCNA4 | 14928 | 4489 | 0.013 | -0.1768 | No | ||
| 11 | CACNG1 | 20177 | 4575 | 0.013 | -0.1787 | No | ||
| 12 | CACNA1C | 1158 1166 24464 4463 8674 | 4965 | 0.011 | -0.1974 | No | ||
| 13 | KCNE1 | 9206 22537 4942 | 6082 | 0.006 | -0.2562 | No | ||
| 14 | KCNIP2 | 23655 3725 | 6132 | 0.006 | -0.2576 | No | ||
| 15 | CACNA1A | 4461 | 6462 | 0.005 | -0.2742 | No | ||
| 16 | CACNA1E | 4465 | 6521 | 0.005 | -0.2763 | No | ||
| 17 | SCN9A | 14571 | 6637 | 0.004 | -0.2816 | No | ||
| 18 | KCNH3 | 9207 | 6768 | 0.004 | -0.2877 | No | ||
| 19 | CACNB3 | 22373 8679 | 7128 | 0.003 | -0.3063 | No | ||
| 20 | SCN1B | 1148 17872 | 7207 | 0.003 | -0.3099 | No | ||
| 21 | KCNA3 | 15460 | 7821 | 0.002 | -0.3426 | No | ||
| 22 | CACNG5 | 1339 20175 | 7894 | 0.002 | -0.3461 | No | ||
| 23 | KCND3 | 15464 | 7903 | 0.002 | -0.3462 | No | ||
| 24 | KCNH1 | 4074 14008 | 7945 | 0.001 | -0.3481 | No | ||
| 25 | KCNS3 | 21109 | 8160 | 0.001 | -0.3594 | No | ||
| 26 | PKD2 | 16772 | 8239 | 0.001 | -0.3634 | No | ||
| 27 | KCNC4 | 15206 1894 | 8329 | 0.001 | -0.3681 | No | ||
| 28 | CACNA1H | 23077 | 8432 | 0.000 | -0.3735 | No | ||
| 29 | CACNB2 | 4466 8678 | 8470 | 0.000 | -0.3754 | No | ||
| 30 | KCNA5 | 9203 | 8590 | 0.000 | -0.3818 | No | ||
| 31 | KCNH2 | 16592 | 8828 | -0.000 | -0.3945 | No | ||
| 32 | CACNA1B | 8673 4462 | 9729 | -0.002 | -0.4425 | No | ||
| 33 | SCN11A | 18968 | 10321 | -0.004 | -0.4735 | No | ||
| 34 | KCNJ5 | 9209 | 10632 | -0.004 | -0.4893 | No | ||
| 35 | KCNC1 | 9204 | 10891 | -0.005 | -0.5021 | No | ||
| 36 | KCNJ3 | 4944 | 10931 | -0.005 | -0.5030 | No | ||
| 37 | KCNJ15 | 4943 9208 | 11064 | -0.006 | -0.5089 | No | ||
| 38 | CACNG4 | 20176 | 11163 | -0.006 | -0.5130 | No | ||
| 39 | CACNB1 | 1443 1304 | 11269 | -0.006 | -0.5173 | No | ||
| 40 | CLCNKA | 15690 | 11323 | -0.006 | -0.5188 | No | ||
| 41 | KCNJ14 | 9988 5586 9989 | 11903 | -0.008 | -0.5482 | No | ||
| 42 | KCNJ4 | 22205 | 12247 | -0.010 | -0.5647 | No | ||
| 43 | NOX1 | 24066 2622 | 12276 | -0.010 | -0.5641 | No | ||
| 44 | SCN4A | 20184 | 12563 | -0.011 | -0.5771 | No | ||
| 45 | SCN7A | 5413 | 12832 | -0.013 | -0.5888 | No | ||
| 46 | CLCNKB | 7105 | 12954 | -0.014 | -0.5924 | No | ||
| 47 | KCNA2 | 15458 | 13523 | -0.018 | -0.6192 | No | ||
| 48 | CACNB4 | 4467 | 13672 | -0.019 | -0.6231 | No | ||
| 49 | CLCN1 | 8744 | 13975 | -0.022 | -0.6346 | No | ||
| 50 | CACNA1F | 12054 24393 | 14249 | -0.026 | -0.6437 | No | ||
| 51 | KCNJ10 | 14047 | 14323 | -0.027 | -0.6419 | No | ||
| 52 | KCNC3 | 4939 9205 | 14363 | -0.028 | -0.6380 | No | ||
| 53 | CACNA1G | 1237 940 20292 | 14502 | -0.030 | -0.6390 | No | ||
| 54 | KCNQ1 | 18001 4948 | 14921 | -0.039 | -0.6531 | No | ||
| 55 | KCNA1 | 9202 | 15707 | -0.065 | -0.6813 | Yes | ||
| 56 | CACNA1S | 14111 | 15766 | -0.068 | -0.6697 | Yes | ||
| 57 | KCNS1 | 14358 | 16201 | -0.092 | -0.6733 | Yes | ||
| 58 | KCNE2 | 22701 | 16631 | -0.125 | -0.6696 | Yes | ||
| 59 | CACNA2D1 | 8676 | 18490 | -0.925 | -0.5706 | Yes | ||
| 60 | KCNK1 | 18415 | 18529 | -1.258 | -0.3021 | Yes | ||
| 61 | CLCN3 | 8745 3782 3893 4524 | 18555 | -1.426 | 0.0033 | Yes |