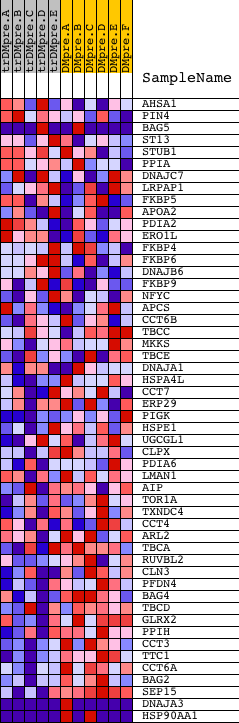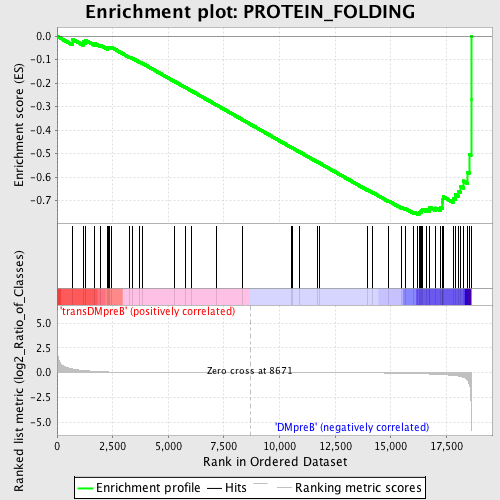
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | PROTEIN_FOLDING |
| Enrichment Score (ES) | -0.75892293 |
| Normalized Enrichment Score (NES) | -1.6223267 |
| Nominal p-value | 0.008791209 |
| FDR q-value | 0.6596641 |
| FWER p-Value | 0.804 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AHSA1 | 21192 | 691 | 0.368 | -0.0129 | No | ||
| 2 | PIN4 | 12817 | 1167 | 0.218 | -0.0240 | No | ||
| 3 | BAG5 | 20981 192 | 1281 | 0.194 | -0.0173 | No | ||
| 4 | ST13 | 12865 | 1693 | 0.130 | -0.0308 | No | ||
| 5 | STUB1 | 23070 | 1954 | 0.097 | -0.0384 | No | ||
| 6 | PPIA | 1284 11188 | 2281 | 0.069 | -0.0514 | No | ||
| 7 | DNAJC7 | 20227 | 2293 | 0.068 | -0.0475 | No | ||
| 8 | LRPAP1 | 5005 | 2356 | 0.065 | -0.0465 | No | ||
| 9 | FKBP5 | 23051 | 2445 | 0.060 | -0.0473 | No | ||
| 10 | APOA2 | 8615 4044 | 3250 | 0.030 | -0.0886 | No | ||
| 11 | PDIA2 | 23062 22604 1702 | 3376 | 0.027 | -0.0936 | No | ||
| 12 | ERO1L | 6930 11924 6929 | 3703 | 0.022 | -0.1097 | No | ||
| 13 | FKBP4 | 4726 | 3817 | 0.020 | -0.1144 | No | ||
| 14 | FKBP6 | 16344 | 5282 | 0.009 | -0.1927 | No | ||
| 15 | DNAJB6 | 6246 3538 | 5769 | 0.007 | -0.2184 | No | ||
| 16 | FKBP9 | 17433 | 6047 | 0.006 | -0.2329 | No | ||
| 17 | NFYC | 9460 | 7166 | 0.003 | -0.2929 | No | ||
| 18 | APCS | 13748 | 8342 | 0.001 | -0.3562 | No | ||
| 19 | CCT6B | 20326 | 10555 | -0.004 | -0.4750 | No | ||
| 20 | TBCC | 23215 | 10575 | -0.004 | -0.4758 | No | ||
| 21 | MKKS | 2946 14416 | 10885 | -0.005 | -0.4921 | No | ||
| 22 | TBCE | 21543 | 11694 | -0.007 | -0.5351 | No | ||
| 23 | DNAJA1 | 4878 16242 2345 | 11771 | -0.008 | -0.5387 | No | ||
| 24 | HSPA4L | 5214 | 13943 | -0.022 | -0.6542 | No | ||
| 25 | CCT7 | 17385 | 14191 | -0.025 | -0.6658 | No | ||
| 26 | ERP29 | 12524 | 14916 | -0.039 | -0.7022 | No | ||
| 27 | PIGK | 1815 6834 11629 | 15475 | -0.055 | -0.7286 | No | ||
| 28 | HSPE1 | 9133 | 15644 | -0.063 | -0.7335 | No | ||
| 29 | UGCGL1 | 6721 | 16020 | -0.081 | -0.7484 | No | ||
| 30 | CLPX | 19403 | 16217 | -0.093 | -0.7527 | Yes | ||
| 31 | PDIA6 | 21308 | 16297 | -0.098 | -0.7505 | Yes | ||
| 32 | LMAN1 | 12867 | 16321 | -0.100 | -0.7451 | Yes | ||
| 33 | AIP | 23955 | 16391 | -0.105 | -0.7419 | Yes | ||
| 34 | TOR1A | 11395 2909 | 16421 | -0.108 | -0.7364 | Yes | ||
| 35 | TXNDC4 | 15886 | 16604 | -0.122 | -0.7381 | Yes | ||
| 36 | CCT4 | 8710 | 16728 | -0.134 | -0.7358 | Yes | ||
| 37 | ARL2 | 23992 | 16743 | -0.135 | -0.7276 | Yes | ||
| 38 | TBCA | 10037 | 17016 | -0.166 | -0.7313 | Yes | ||
| 39 | RUVBL2 | 9766 | 17224 | -0.188 | -0.7300 | Yes | ||
| 40 | CLN3 | 17635 | 17324 | -0.200 | -0.7221 | Yes | ||
| 41 | PFDN4 | 4260 | 17325 | -0.200 | -0.7088 | Yes | ||
| 42 | BAG4 | 7431 | 17334 | -0.201 | -0.6959 | Yes | ||
| 43 | TBCD | 20555 | 17345 | -0.203 | -0.6830 | Yes | ||
| 44 | GLRX2 | 12795 | 17799 | -0.276 | -0.6892 | Yes | ||
| 45 | PPIH | 12287 | 17901 | -0.299 | -0.6748 | Yes | ||
| 46 | CCT3 | 8709 | 18037 | -0.334 | -0.6600 | Yes | ||
| 47 | TTC1 | 7342 | 18139 | -0.380 | -0.6403 | Yes | ||
| 48 | CCT6A | 16689 | 18256 | -0.459 | -0.6162 | Yes | ||
| 49 | BAG2 | 13985 | 18444 | -0.725 | -0.5783 | Yes | ||
| 50 | SEP15 | 13527 1895 15398 | 18524 | -1.196 | -0.5033 | Yes | ||
| 51 | DNAJA3 | 1732 13518 | 18605 | -3.579 | -0.2707 | Yes | ||
| 52 | HSP90AA1 | 4883 4882 9131 | 18608 | -4.096 | 0.0004 | Yes |