Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
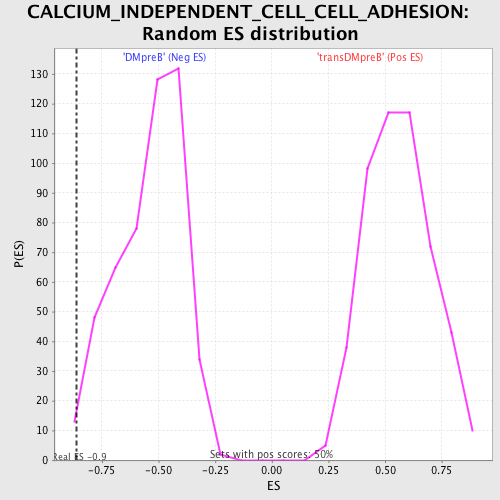
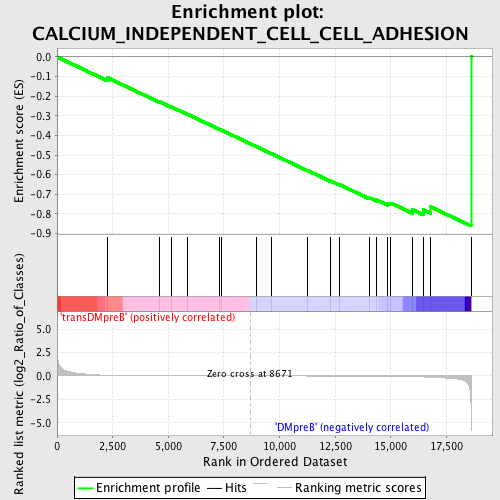
| GeneSet | CALCIUM_INDEPENDENT_CELL_CELL_ADHESION |
| Enrichment Score (ES) | -0.8614118 |
| Normalized Enrichment Score (NES) | -1.6010585 |
| Nominal p-value | 0.014 |
| FDR q-value | 0.60515064 |
| FWER p-Value | 0.919 |

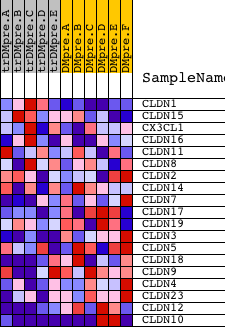
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CLDN1 | 22623 | 2266 | 0.070 | -0.1056 | No | ||
| 2 | CLDN15 | 16667 3572 | 4579 | 0.013 | -0.2269 | No | ||
| 3 | CX3CL1 | 9791 | 5134 | 0.010 | -0.2545 | No | ||
| 4 | CLDN16 | 22802 | 5861 | 0.007 | -0.2920 | No | ||
| 5 | CLDN11 | 15616 | 7276 | 0.003 | -0.3673 | No | ||
| 6 | CLDN8 | 22549 1747 | 7388 | 0.003 | -0.3727 | No | ||
| 7 | CLDN2 | 4527 | 8969 | -0.001 | -0.4575 | No | ||
| 8 | CLDN14 | 12081 | 9622 | -0.002 | -0.4921 | No | ||
| 9 | CLDN7 | 20817 | 11234 | -0.006 | -0.5773 | No | ||
| 10 | CLDN17 | 10757 | 12285 | -0.010 | -0.6315 | No | ||
| 11 | CLDN19 | 16105 472 | 12695 | -0.012 | -0.6507 | No | ||
| 12 | CLDN3 | 16680 | 14022 | -0.023 | -0.7167 | No | ||
| 13 | CLDN5 | 22826 | 14356 | -0.028 | -0.7282 | No | ||
| 14 | CLDN18 | 19028 | 14864 | -0.038 | -0.7467 | No | ||
| 15 | CLDN9 | 24470 | 15001 | -0.041 | -0.7445 | No | ||
| 16 | CLDN4 | 8747 | 15979 | -0.079 | -0.7787 | Yes | ||
| 17 | CLDN23 | 18893 | 16458 | -0.111 | -0.7787 | Yes | ||
| 18 | CLDN12 | 16925 | 16794 | -0.141 | -0.7640 | Yes | ||
| 19 | CLDN10 | 21935 2785 2601 2720 7188 | 18606 | -3.713 | 0.0005 | Yes |