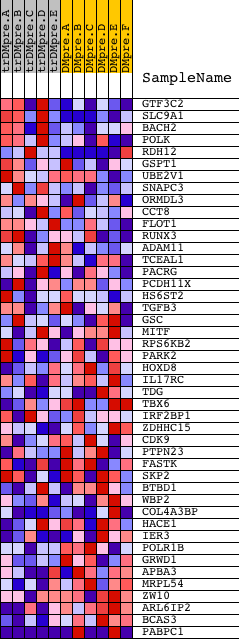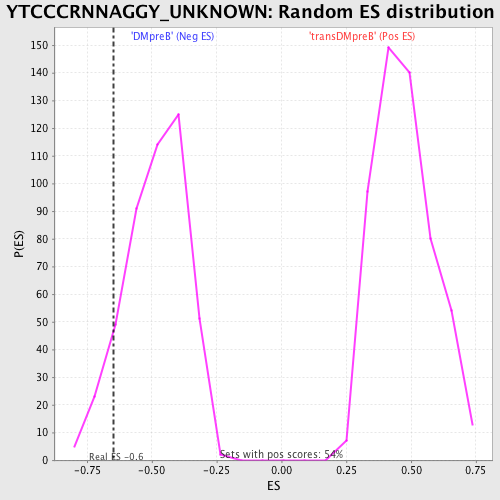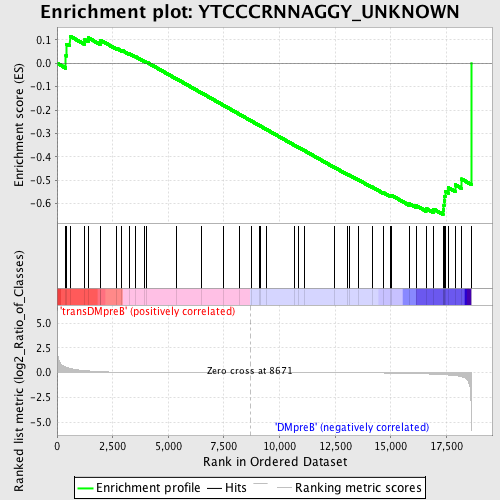
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | YTCCCRNNAGGY_UNKNOWN |
| Enrichment Score (ES) | -0.64601785 |
| Normalized Enrichment Score (NES) | -1.3351074 |
| Nominal p-value | 0.10217391 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GTF3C2 | 7749 | 394 | 0.543 | 0.0321 | No | ||
| 2 | SLC9A1 | 16053 | 425 | 0.520 | 0.0816 | No | ||
| 3 | BACH2 | 8648 | 579 | 0.429 | 0.1156 | No | ||
| 4 | POLK | 21384 | 1246 | 0.203 | 0.0997 | No | ||
| 5 | RDH12 | 21227 | 1405 | 0.174 | 0.1083 | No | ||
| 6 | GSPT1 | 9046 | 1942 | 0.099 | 0.0891 | No | ||
| 7 | UBE2V1 | 12381 | 1952 | 0.098 | 0.0982 | No | ||
| 8 | SNAPC3 | 13397 2456 8085 | 2671 | 0.048 | 0.0642 | No | ||
| 9 | ORMDL3 | 12386 | 2914 | 0.040 | 0.0551 | No | ||
| 10 | CCT8 | 22551 | 3240 | 0.031 | 0.0406 | No | ||
| 11 | FLOT1 | 23249 1613 | 3507 | 0.025 | 0.0287 | No | ||
| 12 | RUNX3 | 8702 | 3943 | 0.019 | 0.0071 | No | ||
| 13 | ADAM11 | 4340 1432 | 4038 | 0.018 | 0.0038 | No | ||
| 14 | TCEAL1 | 24245 | 5365 | 0.009 | -0.0667 | No | ||
| 15 | PACRG | 23127 | 6476 | 0.005 | -0.1260 | No | ||
| 16 | PCDH11X | 2559 | 7492 | 0.002 | -0.1804 | No | ||
| 17 | HS6ST2 | 6939 2646 | 8207 | 0.001 | -0.2188 | No | ||
| 18 | TGFB3 | 10161 | 8719 | -0.000 | -0.2463 | No | ||
| 19 | GSC | 20990 | 9107 | -0.001 | -0.2670 | No | ||
| 20 | MITF | 17349 | 9127 | -0.001 | -0.2680 | No | ||
| 21 | RPS6KB2 | 7198 | 9415 | -0.002 | -0.2833 | No | ||
| 22 | PARK2 | 6944 11939 1499 23385 | 10691 | -0.005 | -0.3515 | No | ||
| 23 | HOXD8 | 9116 | 10854 | -0.005 | -0.3597 | No | ||
| 24 | IL17RC | 17328 1108 | 11124 | -0.006 | -0.3736 | No | ||
| 25 | TDG | 10107 5703 | 12453 | -0.011 | -0.4441 | No | ||
| 26 | TBX6 | 1993 18075 | 13068 | -0.014 | -0.4757 | No | ||
| 27 | IRF2BP1 | 18368 | 13126 | -0.015 | -0.4774 | No | ||
| 28 | ZDHHC15 | 24084 | 13535 | -0.018 | -0.4976 | No | ||
| 29 | CDK9 | 14619 | 14154 | -0.024 | -0.5285 | No | ||
| 30 | PTPN23 | 18988 8357 | 14657 | -0.033 | -0.5523 | No | ||
| 31 | FASTK | 16589 3661 3478 3664 | 15006 | -0.041 | -0.5669 | No | ||
| 32 | SKP2 | 6576 11337 2236 | 15048 | -0.042 | -0.5650 | No | ||
| 33 | BTBD1 | 13519 | 15832 | -0.071 | -0.6002 | No | ||
| 34 | WBP2 | 20146 | 16133 | -0.087 | -0.6078 | No | ||
| 35 | COL4A3BP | 3221 7522 | 16583 | -0.120 | -0.6201 | No | ||
| 36 | HACE1 | 20034 | 16915 | -0.155 | -0.6227 | No | ||
| 37 | IER3 | 23248 | 17349 | -0.204 | -0.6260 | Yes | ||
| 38 | POLR1B | 14857 | 17382 | -0.208 | -0.6073 | Yes | ||
| 39 | GRWD1 | 8317 | 17397 | -0.210 | -0.5874 | Yes | ||
| 40 | APBA3 | 12180 | 17424 | -0.213 | -0.5679 | Yes | ||
| 41 | MRPL54 | 19678 | 17449 | -0.218 | -0.5477 | Yes | ||
| 42 | ZW10 | 3114 19464 | 17581 | -0.236 | -0.5316 | Yes | ||
| 43 | ARL6IP2 | 1496 7090 12093 | 17895 | -0.297 | -0.5193 | Yes | ||
| 44 | BCAS3 | 5314 | 18177 | -0.408 | -0.4943 | Yes | ||
| 45 | PABPC1 | 5219 9522 9523 23572 | 18615 | -5.269 | 0.0001 | Yes |