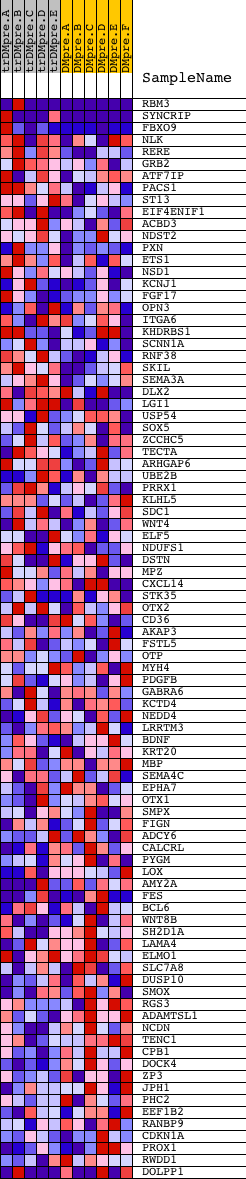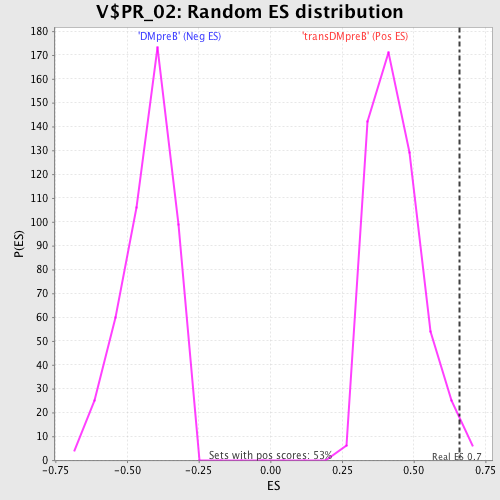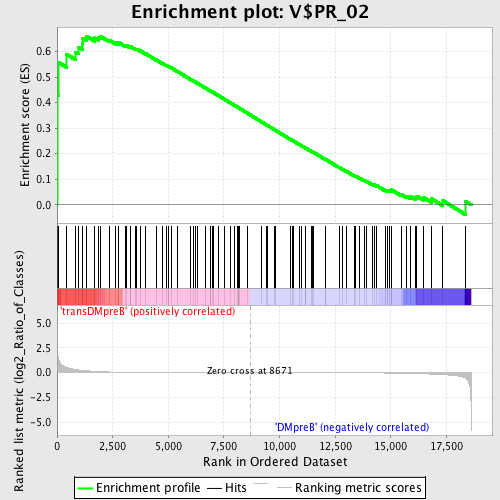
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$PR_02 |
| Enrichment Score (ES) | 0.6602144 |
| Normalized Enrichment Score (NES) | 1.5087575 |
| Nominal p-value | 0.011257036 |
| FDR q-value | 0.2813211 |
| FWER p-Value | 0.957 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RBM3 | 5366 2625 | 2 | 4.390 | 0.4315 | Yes | ||
| 2 | SYNCRIP | 3078 3035 3107 | 80 | 1.313 | 0.5564 | Yes | ||
| 3 | FBXO9 | 19058 3146 | 414 | 0.525 | 0.5901 | Yes | ||
| 4 | NLK | 5179 5178 | 838 | 0.311 | 0.5978 | Yes | ||
| 5 | RERE | 12722 | 970 | 0.270 | 0.6173 | Yes | ||
| 6 | GRB2 | 20149 | 1140 | 0.224 | 0.6302 | Yes | ||
| 7 | ATF7IP | 12028 13038 | 1156 | 0.220 | 0.6510 | Yes | ||
| 8 | PACS1 | 23975 | 1322 | 0.184 | 0.6602 | Yes | ||
| 9 | ST13 | 12865 | 1693 | 0.130 | 0.6530 | No | ||
| 10 | EIF4ENIF1 | 20971 | 1866 | 0.108 | 0.6543 | No | ||
| 11 | ACBD3 | 14025 | 1951 | 0.098 | 0.6594 | No | ||
| 12 | NDST2 | 21906 | 2340 | 0.066 | 0.6449 | No | ||
| 13 | PXN | 5339 3573 | 2632 | 0.049 | 0.6340 | No | ||
| 14 | ETS1 | 10715 6230 3135 | 2762 | 0.044 | 0.6314 | No | ||
| 15 | NSD1 | 2134 5197 | 2777 | 0.044 | 0.6350 | No | ||
| 16 | KCNJ1 | 3121 12110 | 3090 | 0.034 | 0.6215 | No | ||
| 17 | FGF17 | 21757 | 3120 | 0.034 | 0.6232 | No | ||
| 18 | OPN3 | 13740 | 3285 | 0.029 | 0.6173 | No | ||
| 19 | ITGA6 | 4930 | 3320 | 0.029 | 0.6182 | No | ||
| 20 | KHDRBS1 | 5405 9778 | 3543 | 0.024 | 0.6086 | No | ||
| 21 | SCNN1A | 17278 | 3582 | 0.024 | 0.6089 | No | ||
| 22 | RNF38 | 7862 2328 13115 | 3736 | 0.021 | 0.6027 | No | ||
| 23 | SKIL | 15617 | 3981 | 0.018 | 0.5914 | No | ||
| 24 | SEMA3A | 16919 | 4448 | 0.014 | 0.5676 | No | ||
| 25 | DLX2 | 14561 353 | 4740 | 0.012 | 0.5530 | No | ||
| 26 | LGI1 | 23867 | 4903 | 0.011 | 0.5454 | No | ||
| 27 | USP54 | 21910 | 5002 | 0.010 | 0.5411 | No | ||
| 28 | SOX5 | 9850 1044 5480 16937 | 5016 | 0.010 | 0.5414 | No | ||
| 29 | ZCCHC5 | 24081 | 5120 | 0.010 | 0.5368 | No | ||
| 30 | TECTA | 19159 | 5432 | 0.008 | 0.5208 | No | ||
| 31 | ARHGAP6 | 24207 | 5985 | 0.006 | 0.4916 | No | ||
| 32 | UBE2B | 10245 | 6118 | 0.006 | 0.4851 | No | ||
| 33 | PRRX1 | 9595 5271 | 6205 | 0.006 | 0.4810 | No | ||
| 34 | KLHL5 | 3631 7753 | 6293 | 0.005 | 0.4768 | No | ||
| 35 | SDC1 | 5560 9953 | 6649 | 0.004 | 0.4581 | No | ||
| 36 | WNT4 | 16025 | 6898 | 0.004 | 0.4451 | No | ||
| 37 | ELF5 | 14936 2913 | 6912 | 0.004 | 0.4448 | No | ||
| 38 | NDUFS1 | 13940 | 6973 | 0.004 | 0.4419 | No | ||
| 39 | DSTN | 14823 | 7021 | 0.003 | 0.4397 | No | ||
| 40 | MPZ | 9408 | 7245 | 0.003 | 0.4279 | No | ||
| 41 | CXCL14 | 21445 7165 | 7545 | 0.002 | 0.4120 | No | ||
| 42 | STK35 | 14851 | 7803 | 0.002 | 0.3983 | No | ||
| 43 | OTX2 | 9519 | 7990 | 0.001 | 0.3884 | No | ||
| 44 | CD36 | 8712 | 8104 | 0.001 | 0.3824 | No | ||
| 45 | AKAP3 | 8566 | 8152 | 0.001 | 0.3800 | No | ||
| 46 | FSTL5 | 10018 | 8168 | 0.001 | 0.3793 | No | ||
| 47 | OTP | 21582 | 8180 | 0.001 | 0.3788 | No | ||
| 48 | MYH4 | 20837 | 8557 | 0.000 | 0.3585 | No | ||
| 49 | PDGFB | 22199 2311 | 9170 | -0.001 | 0.3256 | No | ||
| 50 | GABRA6 | 20491 | 9428 | -0.002 | 0.3119 | No | ||
| 51 | KCTD4 | 21956 | 9466 | -0.002 | 0.3101 | No | ||
| 52 | NEDD4 | 19384 3101 | 9763 | -0.002 | 0.2943 | No | ||
| 53 | LRRTM3 | 19744 | 9799 | -0.002 | 0.2927 | No | ||
| 54 | BDNF | 14926 2797 | 10471 | -0.004 | 0.2568 | No | ||
| 55 | KRT20 | 12410 | 10561 | -0.004 | 0.2525 | No | ||
| 56 | MBP | 23502 | 10641 | -0.005 | 0.2487 | No | ||
| 57 | SEMA4C | 9799 | 10872 | -0.005 | 0.2367 | No | ||
| 58 | EPHA7 | 8909 4674 | 10963 | -0.005 | 0.2324 | No | ||
| 59 | OTX1 | 20517 | 11148 | -0.006 | 0.2230 | No | ||
| 60 | SMPX | 24220 | 11161 | -0.006 | 0.2230 | No | ||
| 61 | FIGN | 14575 | 11442 | -0.007 | 0.2085 | No | ||
| 62 | ADCY6 | 22139 2283 8551 | 11485 | -0.007 | 0.2069 | No | ||
| 63 | CALCRL | 12041 | 11503 | -0.007 | 0.2067 | No | ||
| 64 | PYGM | 23792 | 12049 | -0.009 | 0.1781 | No | ||
| 65 | LOX | 23438 | 12074 | -0.009 | 0.1777 | No | ||
| 66 | AMY2A | 8587 | 12674 | -0.012 | 0.1465 | No | ||
| 67 | FES | 17779 3949 | 12817 | -0.013 | 0.1401 | No | ||
| 68 | BCL6 | 22624 | 13018 | -0.014 | 0.1307 | No | ||
| 69 | WNT8B | 23841 | 13379 | -0.016 | 0.1128 | No | ||
| 70 | SH2D1A | 9810 | 13412 | -0.017 | 0.1127 | No | ||
| 71 | LAMA4 | 20054 | 13611 | -0.019 | 0.1039 | No | ||
| 72 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13808 | -0.021 | 0.0953 | No | ||
| 73 | SLC7A8 | 3611 21833 | 13909 | -0.022 | 0.0920 | No | ||
| 74 | DUSP10 | 4003 14016 | 14161 | -0.024 | 0.0809 | No | ||
| 75 | SMOX | 10455 2940 2737 2952 | 14261 | -0.026 | 0.0781 | No | ||
| 76 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 14351 | -0.027 | 0.0760 | No | ||
| 77 | ADAMTSL1 | 16181 | 14749 | -0.035 | 0.0580 | No | ||
| 78 | NCDN | 15756 2487 6471 | 14856 | -0.038 | 0.0560 | No | ||
| 79 | TENC1 | 22349 9927 | 14944 | -0.040 | 0.0552 | No | ||
| 80 | CPB1 | 175 | 15013 | -0.041 | 0.0556 | No | ||
| 81 | DOCK4 | 10705 | 15019 | -0.041 | 0.0594 | No | ||
| 82 | ZP3 | 3509 | 15470 | -0.055 | 0.0405 | No | ||
| 83 | JPH1 | 13994 | 15713 | -0.066 | 0.0339 | No | ||
| 84 | PHC2 | 16075 | 15901 | -0.075 | 0.0312 | No | ||
| 85 | EEF1B2 | 4131 12063 | 16101 | -0.085 | 0.0289 | No | ||
| 86 | RANBP9 | 21480 | 16173 | -0.090 | 0.0339 | No | ||
| 87 | CDKN1A | 4511 8729 | 16471 | -0.111 | 0.0288 | No | ||
| 88 | PROX1 | 9623 | 16841 | -0.147 | 0.0234 | No | ||
| 89 | RWDD1 | 7306 | 17343 | -0.203 | 0.0163 | No | ||
| 90 | DOLPP1 | 2682 15051 | 18347 | -0.533 | 0.0145 | No |