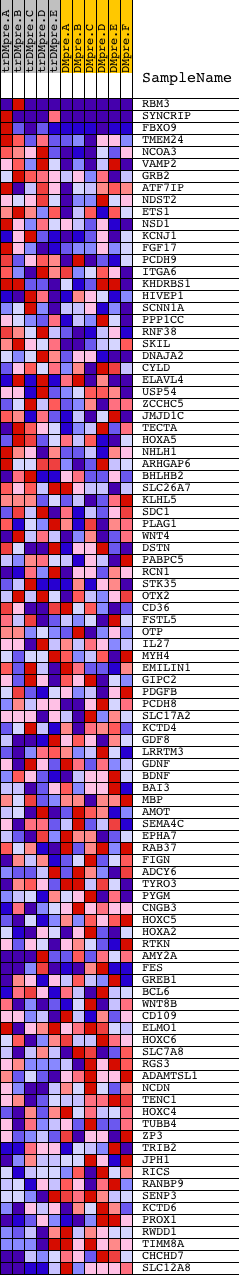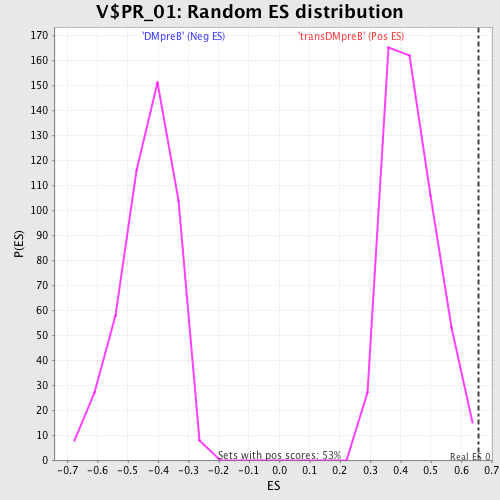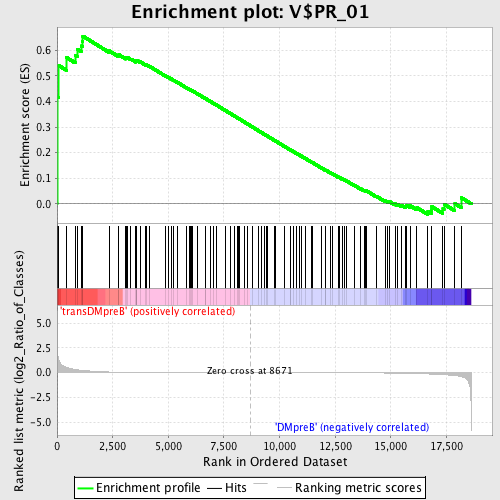
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$PR_01 |
| Enrichment Score (ES) | 0.6558497 |
| Normalized Enrichment Score (NES) | 1.5119535 |
| Nominal p-value | 0.0018939395 |
| FDR q-value | 0.31270862 |
| FWER p-Value | 0.948 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RBM3 | 5366 2625 | 2 | 4.390 | 0.4193 | Yes | ||
| 2 | SYNCRIP | 3078 3035 3107 | 80 | 1.313 | 0.5405 | Yes | ||
| 3 | FBXO9 | 19058 3146 | 414 | 0.525 | 0.5727 | Yes | ||
| 4 | TMEM24 | 19150 | 814 | 0.318 | 0.5816 | Yes | ||
| 5 | NCOA3 | 5156 | 919 | 0.288 | 0.6035 | Yes | ||
| 6 | VAMP2 | 20826 | 1082 | 0.237 | 0.6173 | Yes | ||
| 7 | GRB2 | 20149 | 1140 | 0.224 | 0.6356 | Yes | ||
| 8 | ATF7IP | 12028 13038 | 1156 | 0.220 | 0.6558 | Yes | ||
| 9 | NDST2 | 21906 | 2340 | 0.066 | 0.5982 | No | ||
| 10 | ETS1 | 10715 6230 3135 | 2762 | 0.044 | 0.5797 | No | ||
| 11 | NSD1 | 2134 5197 | 2777 | 0.044 | 0.5832 | No | ||
| 12 | KCNJ1 | 3121 12110 | 3090 | 0.034 | 0.5696 | No | ||
| 13 | FGF17 | 21757 | 3120 | 0.034 | 0.5712 | No | ||
| 14 | PCDH9 | 21736 | 3168 | 0.032 | 0.5718 | No | ||
| 15 | ITGA6 | 4930 | 3320 | 0.029 | 0.5664 | No | ||
| 16 | KHDRBS1 | 5405 9778 | 3543 | 0.024 | 0.5567 | No | ||
| 17 | HIVEP1 | 8486 | 3551 | 0.024 | 0.5586 | No | ||
| 18 | SCNN1A | 17278 | 3582 | 0.024 | 0.5592 | No | ||
| 19 | PPP1CC | 9609 5283 | 3588 | 0.024 | 0.5612 | No | ||
| 20 | RNF38 | 7862 2328 13115 | 3736 | 0.021 | 0.5553 | No | ||
| 21 | SKIL | 15617 | 3981 | 0.018 | 0.5439 | No | ||
| 22 | DNAJA2 | 12133 18806 | 4012 | 0.018 | 0.5440 | No | ||
| 23 | CYLD | 18532 | 4163 | 0.016 | 0.5374 | No | ||
| 24 | ELAVL4 | 15805 4889 9137 | 4871 | 0.011 | 0.5003 | No | ||
| 25 | USP54 | 21910 | 5002 | 0.010 | 0.4943 | No | ||
| 26 | ZCCHC5 | 24081 | 5120 | 0.010 | 0.4889 | No | ||
| 27 | JMJD1C | 11531 19996 | 5228 | 0.009 | 0.4840 | No | ||
| 28 | TECTA | 19159 | 5432 | 0.008 | 0.4738 | No | ||
| 29 | HOXA5 | 17149 | 5806 | 0.007 | 0.4544 | No | ||
| 30 | NHLH1 | 13754 | 5950 | 0.006 | 0.4472 | No | ||
| 31 | ARHGAP6 | 24207 | 5985 | 0.006 | 0.4460 | No | ||
| 32 | BHLHB2 | 17340 | 6018 | 0.006 | 0.4449 | No | ||
| 33 | SLC26A7 | 5538 2372 | 6106 | 0.006 | 0.4408 | No | ||
| 34 | KLHL5 | 3631 7753 | 6293 | 0.005 | 0.4312 | No | ||
| 35 | SDC1 | 5560 9953 | 6649 | 0.004 | 0.4125 | No | ||
| 36 | PLAG1 | 7148 15949 12161 | 6690 | 0.004 | 0.4107 | No | ||
| 37 | WNT4 | 16025 | 6898 | 0.004 | 0.3999 | No | ||
| 38 | DSTN | 14823 | 7021 | 0.003 | 0.3937 | No | ||
| 39 | PABPC5 | 24077 | 7155 | 0.003 | 0.3868 | No | ||
| 40 | RCN1 | 9710 | 7577 | 0.002 | 0.3643 | No | ||
| 41 | STK35 | 14851 | 7803 | 0.002 | 0.3523 | No | ||
| 42 | OTX2 | 9519 | 7990 | 0.001 | 0.3424 | No | ||
| 43 | CD36 | 8712 | 8104 | 0.001 | 0.3364 | No | ||
| 44 | FSTL5 | 10018 | 8168 | 0.001 | 0.3331 | No | ||
| 45 | OTP | 21582 | 8180 | 0.001 | 0.3326 | No | ||
| 46 | IL27 | 17636 | 8418 | 0.001 | 0.3198 | No | ||
| 47 | MYH4 | 20837 | 8557 | 0.000 | 0.3124 | No | ||
| 48 | EMILIN1 | 16891 | 8786 | -0.000 | 0.3001 | No | ||
| 49 | GIPC2 | 15137 | 9072 | -0.001 | 0.2848 | No | ||
| 50 | PDGFB | 22199 2311 | 9170 | -0.001 | 0.2796 | No | ||
| 51 | PCDH8 | 21739 3017 | 9303 | -0.001 | 0.2726 | No | ||
| 52 | SLC17A2 | 10163 5744 | 9420 | -0.002 | 0.2665 | No | ||
| 53 | KCTD4 | 21956 | 9466 | -0.002 | 0.2643 | No | ||
| 54 | GDF8 | 9411 | 9791 | -0.002 | 0.2470 | No | ||
| 55 | LRRTM3 | 19744 | 9799 | -0.002 | 0.2469 | No | ||
| 56 | GDNF | 22514 2275 | 10216 | -0.003 | 0.2247 | No | ||
| 57 | BDNF | 14926 2797 | 10471 | -0.004 | 0.2114 | No | ||
| 58 | BAI3 | 5580 | 10501 | -0.004 | 0.2102 | No | ||
| 59 | MBP | 23502 | 10641 | -0.005 | 0.2032 | No | ||
| 60 | AMOT | 11341 24037 6587 | 10754 | -0.005 | 0.1976 | No | ||
| 61 | SEMA4C | 9799 | 10872 | -0.005 | 0.1917 | No | ||
| 62 | EPHA7 | 8909 4674 | 10963 | -0.005 | 0.1874 | No | ||
| 63 | RAB37 | 20602 | 11166 | -0.006 | 0.1770 | No | ||
| 64 | FIGN | 14575 | 11442 | -0.007 | 0.1628 | No | ||
| 65 | ADCY6 | 22139 2283 8551 | 11485 | -0.007 | 0.1612 | No | ||
| 66 | TYRO3 | 5811 | 11880 | -0.008 | 0.1407 | No | ||
| 67 | PYGM | 23792 | 12049 | -0.009 | 0.1325 | No | ||
| 68 | CNGB3 | 16267 | 12073 | -0.009 | 0.1321 | No | ||
| 69 | HOXC5 | 22340 | 12284 | -0.010 | 0.1217 | No | ||
| 70 | HOXA2 | 17151 | 12362 | -0.010 | 0.1185 | No | ||
| 71 | RTKN | 17394 1156 | 12629 | -0.012 | 0.1052 | No | ||
| 72 | AMY2A | 8587 | 12674 | -0.012 | 0.1040 | No | ||
| 73 | FES | 17779 3949 | 12817 | -0.013 | 0.0975 | No | ||
| 74 | GREB1 | 6490 2152 | 12908 | -0.013 | 0.0939 | No | ||
| 75 | BCL6 | 22624 | 13018 | -0.014 | 0.0894 | No | ||
| 76 | WNT8B | 23841 | 13379 | -0.016 | 0.0715 | No | ||
| 77 | CD109 | 6174 10656 19369 | 13645 | -0.019 | 0.0590 | No | ||
| 78 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13808 | -0.021 | 0.0522 | No | ||
| 79 | HOXC6 | 22339 2302 | 13875 | -0.021 | 0.0506 | No | ||
| 80 | SLC7A8 | 3611 21833 | 13909 | -0.022 | 0.0509 | No | ||
| 81 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 14351 | -0.027 | 0.0297 | No | ||
| 82 | ADAMTSL1 | 16181 | 14749 | -0.035 | 0.0116 | No | ||
| 83 | NCDN | 15756 2487 6471 | 14856 | -0.038 | 0.0095 | No | ||
| 84 | TENC1 | 22349 9927 | 14944 | -0.040 | 0.0086 | No | ||
| 85 | HOXC4 | 4864 | 15189 | -0.046 | -0.0002 | No | ||
| 86 | TUBB4 | 22917 | 15320 | -0.050 | -0.0024 | No | ||
| 87 | ZP3 | 3509 | 15470 | -0.055 | -0.0052 | No | ||
| 88 | TRIB2 | 21102 | 15668 | -0.064 | -0.0097 | No | ||
| 89 | JPH1 | 13994 | 15713 | -0.066 | -0.0058 | No | ||
| 90 | RICS | 19523 | 15861 | -0.073 | -0.0068 | No | ||
| 91 | RANBP9 | 21480 | 16173 | -0.090 | -0.0150 | No | ||
| 92 | SENP3 | 20387 | 16666 | -0.128 | -0.0293 | No | ||
| 93 | KCTD6 | 22098 | 16820 | -0.144 | -0.0237 | No | ||
| 94 | PROX1 | 9623 | 16841 | -0.147 | -0.0108 | No | ||
| 95 | RWDD1 | 7306 | 17343 | -0.203 | -0.0185 | No | ||
| 96 | TIMM8A | 24062 | 17394 | -0.209 | -0.0012 | No | ||
| 97 | CHCHD7 | 16280 | 17881 | -0.294 | 0.0006 | No | ||
| 98 | SLC12A8 | 5059 | 18178 | -0.408 | 0.0237 | No |