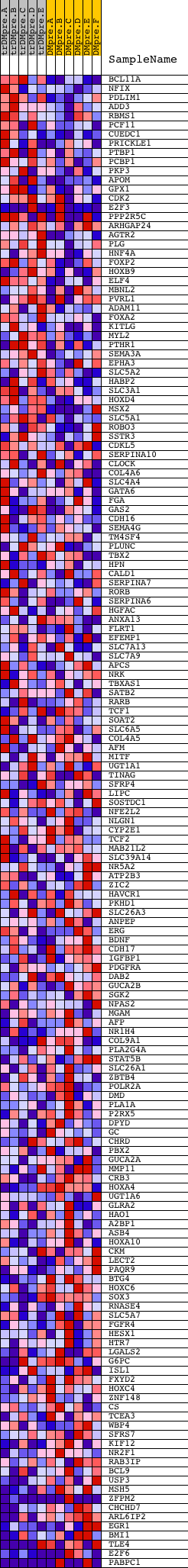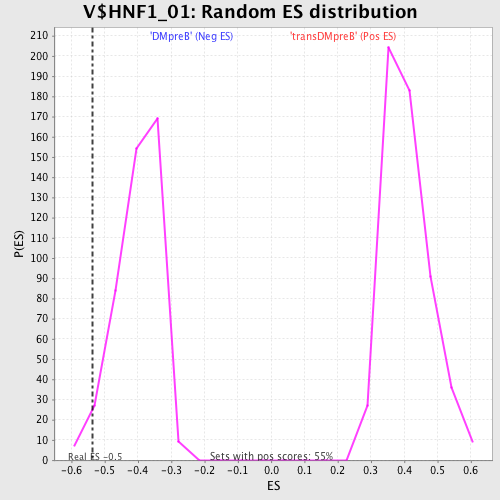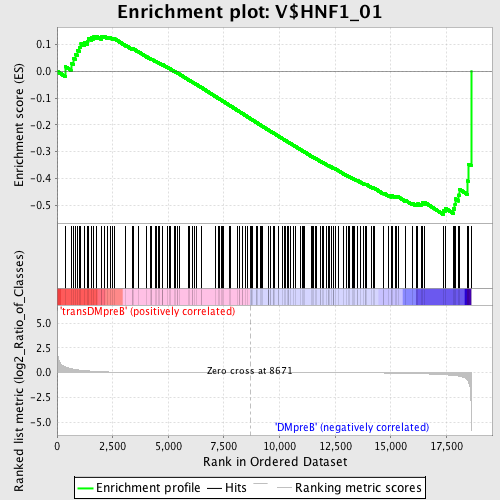
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$HNF1_01 |
| Enrichment Score (ES) | -0.53538084 |
| Normalized Enrichment Score (NES) | -1.3413682 |
| Nominal p-value | 0.033333335 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BCL11A | 4691 | 380 | 0.557 | 0.0169 | No | ||
| 2 | NFIX | 9458 | 630 | 0.396 | 0.0300 | No | ||
| 3 | PDLIM1 | 7023 | 737 | 0.346 | 0.0475 | No | ||
| 4 | ADD3 | 23812 3694 | 823 | 0.317 | 0.0642 | No | ||
| 5 | RBMS1 | 14580 | 923 | 0.287 | 0.0781 | No | ||
| 6 | PCF11 | 121 | 1016 | 0.256 | 0.0903 | No | ||
| 7 | CUEDC1 | 8343 | 1070 | 0.242 | 0.1037 | No | ||
| 8 | PRICKLE1 | 8379 | 1226 | 0.206 | 0.1092 | No | ||
| 9 | PTBP1 | 5303 | 1380 | 0.176 | 0.1128 | No | ||
| 10 | PCBP1 | 17085 | 1389 | 0.175 | 0.1241 | No | ||
| 11 | PKP3 | 18016 | 1539 | 0.153 | 0.1263 | No | ||
| 12 | APOM | 23007 | 1651 | 0.136 | 0.1294 | No | ||
| 13 | GPX1 | 19310 | 1767 | 0.122 | 0.1314 | No | ||
| 14 | CDK2 | 3438 3373 19592 3322 | 1996 | 0.093 | 0.1253 | No | ||
| 15 | E2F3 | 21500 8874 | 2002 | 0.092 | 0.1312 | No | ||
| 16 | PPP2R5C | 6522 2147 | 2122 | 0.082 | 0.1302 | No | ||
| 17 | ARHGAP24 | 16778 3655 | 2280 | 0.069 | 0.1263 | No | ||
| 18 | AGTR2 | 24365 | 2380 | 0.063 | 0.1252 | No | ||
| 19 | PLG | 1593 23383 | 2488 | 0.057 | 0.1233 | No | ||
| 20 | HNF4A | 14746 | 2557 | 0.053 | 0.1231 | No | ||
| 21 | FOXP2 | 4312 17523 8523 | 3091 | 0.034 | 0.0965 | No | ||
| 22 | HOXB9 | 20689 | 3366 | 0.028 | 0.0835 | No | ||
| 23 | ELF4 | 24162 | 3391 | 0.027 | 0.0840 | No | ||
| 24 | MBNL2 | 21932 | 3427 | 0.026 | 0.0839 | No | ||
| 25 | PVRL1 | 3006 19482 12211 7192 | 3636 | 0.023 | 0.0742 | No | ||
| 26 | ADAM11 | 4340 1432 | 4038 | 0.018 | 0.0536 | No | ||
| 27 | FOXA2 | 14405 2889 | 4181 | 0.016 | 0.0470 | No | ||
| 28 | KITLG | 19889 3342 | 4230 | 0.016 | 0.0455 | No | ||
| 29 | MYL2 | 16713 | 4258 | 0.015 | 0.0450 | No | ||
| 30 | PTHR1 | 5318 18985 | 4407 | 0.014 | 0.0379 | No | ||
| 31 | SEMA3A | 16919 | 4448 | 0.014 | 0.0367 | No | ||
| 32 | EPHA3 | 22563 1643 | 4534 | 0.013 | 0.0330 | No | ||
| 33 | SLC5A2 | 18057 10905 | 4607 | 0.013 | 0.0299 | No | ||
| 34 | HABP2 | 10364 23825 | 4734 | 0.012 | 0.0239 | No | ||
| 35 | SLC3A1 | 23145 | 4757 | 0.012 | 0.0235 | No | ||
| 36 | HOXD4 | 4866 | 4758 | 0.012 | 0.0243 | No | ||
| 37 | MSX2 | 21461 | 4979 | 0.010 | 0.0131 | No | ||
| 38 | SLC5A1 | 5455 | 5039 | 0.010 | 0.0105 | No | ||
| 39 | ROBO3 | 9705 | 5076 | 0.010 | 0.0093 | No | ||
| 40 | SSTR3 | 22218 | 5281 | 0.009 | -0.0012 | No | ||
| 41 | CDKL5 | 24019 | 5340 | 0.009 | -0.0037 | No | ||
| 42 | SERPINA10 | 10151 | 5390 | 0.009 | -0.0058 | No | ||
| 43 | CLOCK | 8749 16507 4530 | 5424 | 0.008 | -0.0071 | No | ||
| 44 | COL4A6 | 8248 2552 | 5496 | 0.008 | -0.0104 | No | ||
| 45 | SLC4A4 | 16807 12035 7036 | 5922 | 0.007 | -0.0329 | No | ||
| 46 | GATA6 | 96 | 5946 | 0.006 | -0.0338 | No | ||
| 47 | FGA | 1780 15566 | 6094 | 0.006 | -0.0413 | No | ||
| 48 | GAS2 | 4753 | 6097 | 0.006 | -0.0410 | No | ||
| 49 | CDH16 | 4507 | 6169 | 0.006 | -0.0445 | No | ||
| 50 | SEMA4G | 11182 | 6276 | 0.005 | -0.0499 | No | ||
| 51 | TM4SF4 | 15588 | 6490 | 0.005 | -0.0611 | No | ||
| 52 | PLUNC | 9593 | 6501 | 0.005 | -0.0613 | No | ||
| 53 | TBX2 | 20720 | 7118 | 0.003 | -0.0944 | No | ||
| 54 | HPN | 17873 4087 | 7242 | 0.003 | -0.1009 | No | ||
| 55 | CALD1 | 4273 8463 | 7286 | 0.003 | -0.1030 | No | ||
| 56 | SERPINA7 | 6856 | 7379 | 0.003 | -0.1079 | No | ||
| 57 | RORB | 10356 | 7448 | 0.003 | -0.1114 | No | ||
| 58 | SERPINA6 | 20993 | 7465 | 0.002 | -0.1121 | No | ||
| 59 | HGFAC | 16871 | 7767 | 0.002 | -0.1283 | No | ||
| 60 | ANXA13 | 22287 | 7791 | 0.002 | -0.1294 | No | ||
| 61 | FLRT1 | 11852 | 8115 | 0.001 | -0.1468 | No | ||
| 62 | EFEMP1 | 20937 | 8175 | 0.001 | -0.1500 | No | ||
| 63 | SLC7A13 | 16266 | 8195 | 0.001 | -0.1509 | No | ||
| 64 | SLC7A9 | 18291 1766 | 8327 | 0.001 | -0.1580 | No | ||
| 65 | APCS | 13748 | 8342 | 0.001 | -0.1587 | No | ||
| 66 | NRK | 6559 | 8480 | 0.000 | -0.1661 | No | ||
| 67 | TBXAS1 | 17480 1105 | 8565 | 0.000 | -0.1706 | No | ||
| 68 | SATB2 | 5604 | 8702 | -0.000 | -0.1780 | No | ||
| 69 | RARB | 3011 21915 | 8748 | -0.000 | -0.1804 | No | ||
| 70 | TCF1 | 16416 | 8752 | -0.000 | -0.1806 | No | ||
| 71 | SOAT2 | 22348 10302 | 8787 | -0.000 | -0.1824 | No | ||
| 72 | SLC6A5 | 18230 | 8957 | -0.001 | -0.1915 | No | ||
| 73 | COL4A5 | 4543 | 8965 | -0.001 | -0.1918 | No | ||
| 74 | AFM | 16803 | 8993 | -0.001 | -0.1933 | No | ||
| 75 | MITF | 17349 | 9127 | -0.001 | -0.2004 | No | ||
| 76 | UGT1A1 | 11851 | 9203 | -0.001 | -0.2044 | No | ||
| 77 | TINAG | 3141 19060 | 9253 | -0.001 | -0.2070 | No | ||
| 78 | SFRP4 | 9806 | 9504 | -0.002 | -0.2204 | No | ||
| 79 | LIPC | 19065 | 9514 | -0.002 | -0.2208 | No | ||
| 80 | SOSTDC1 | 21286 | 9516 | -0.002 | -0.2207 | No | ||
| 81 | NFE2L2 | 2898 14557 | 9522 | -0.002 | -0.2208 | No | ||
| 82 | NLGN1 | 15368 | 9575 | -0.002 | -0.2235 | No | ||
| 83 | CYP2E1 | 18025 | 9584 | -0.002 | -0.2238 | No | ||
| 84 | TCF2 | 1395 20727 | 9721 | -0.002 | -0.2310 | No | ||
| 85 | MAB21L2 | 15305 | 9738 | -0.002 | -0.2317 | No | ||
| 86 | SLC39A14 | 3517 5611 21762 | 9775 | -0.002 | -0.2335 | No | ||
| 87 | NR5A2 | 4105 13817 | 9940 | -0.003 | -0.2422 | No | ||
| 88 | ATP2B3 | 24308 11557 | 9965 | -0.003 | -0.2433 | No | ||
| 89 | ZIC2 | 21927 | 10130 | -0.003 | -0.2520 | No | ||
| 90 | HAVCR1 | 9360 | 10235 | -0.004 | -0.2574 | No | ||
| 91 | PKHD1 | 6292 4106 | 10256 | -0.004 | -0.2582 | No | ||
| 92 | SLC26A3 | 21298 | 10368 | -0.004 | -0.2640 | No | ||
| 93 | ANPEP | 17784 | 10386 | -0.004 | -0.2647 | No | ||
| 94 | ERG | 1686 8915 | 10399 | -0.004 | -0.2651 | No | ||
| 95 | BDNF | 14926 2797 | 10471 | -0.004 | -0.2686 | No | ||
| 96 | CDH17 | 16273 | 10635 | -0.005 | -0.2772 | No | ||
| 97 | IGFBP1 | 20948 | 10732 | -0.005 | -0.2820 | No | ||
| 98 | PDGFRA | 16824 | 10921 | -0.005 | -0.2919 | No | ||
| 99 | DAB2 | 8835 | 11020 | -0.005 | -0.2968 | No | ||
| 100 | GUCA2B | 15773 | 11071 | -0.006 | -0.2992 | No | ||
| 101 | SGK2 | 14750 | 11138 | -0.006 | -0.3023 | No | ||
| 102 | NPAS2 | 5186 | 11428 | -0.007 | -0.3176 | No | ||
| 103 | MGAM | 17476 | 11484 | -0.007 | -0.3201 | No | ||
| 104 | AFP | 16805 | 11530 | -0.007 | -0.3221 | No | ||
| 105 | NR1H4 | 19648 | 11621 | -0.007 | -0.3265 | No | ||
| 106 | COL9A1 | 3950 4549 | 11660 | -0.007 | -0.3280 | No | ||
| 107 | PLA2G4A | 13809 | 11823 | -0.008 | -0.3363 | No | ||
| 108 | STAT5B | 20222 | 11851 | -0.008 | -0.3372 | No | ||
| 109 | SLC26A1 | 16446 | 11915 | -0.008 | -0.3400 | No | ||
| 110 | ZBTB4 | 13267 | 11975 | -0.008 | -0.3427 | No | ||
| 111 | POLR2A | 5394 | 12103 | -0.009 | -0.3489 | No | ||
| 112 | DMD | 24295 2647 | 12181 | -0.009 | -0.3525 | No | ||
| 113 | PLA1A | 1737 22594 | 12215 | -0.010 | -0.3536 | No | ||
| 114 | P2RX5 | 1370 20790 | 12242 | -0.010 | -0.3544 | No | ||
| 115 | DPYD | 15437 | 12310 | -0.010 | -0.3574 | No | ||
| 116 | GC | 16486 | 12402 | -0.010 | -0.3616 | No | ||
| 117 | CHRD | 22817 | 12438 | -0.011 | -0.3628 | No | ||
| 118 | PBX2 | 1614 23279 | 12440 | -0.011 | -0.3621 | No | ||
| 119 | GUCA2A | 16104 | 12494 | -0.011 | -0.3643 | No | ||
| 120 | MMP11 | 19727 | 12634 | -0.012 | -0.3710 | No | ||
| 121 | CRB3 | 23176 | 12894 | -0.013 | -0.3842 | No | ||
| 122 | HOXA4 | 17148 | 12989 | -0.014 | -0.3884 | No | ||
| 123 | UGT1A6 | 3969 4079 6911 13591 | 13108 | -0.014 | -0.3938 | No | ||
| 124 | GLRA2 | 24008 | 13144 | -0.015 | -0.3947 | No | ||
| 125 | HAO1 | 14418 | 13259 | -0.016 | -0.3998 | No | ||
| 126 | A2BP1 | 11212 1652 1689 | 13329 | -0.016 | -0.4025 | No | ||
| 127 | ASB4 | 990 17529 | 13355 | -0.016 | -0.4028 | No | ||
| 128 | HOXA10 | 17145 989 | 13494 | -0.017 | -0.4091 | No | ||
| 129 | CKM | 18362 | 13495 | -0.017 | -0.4079 | No | ||
| 130 | LECT2 | 21443 | 13628 | -0.019 | -0.4138 | No | ||
| 131 | PAQR9 | 19351 | 13788 | -0.020 | -0.4210 | No | ||
| 132 | BTG4 | 19456 | 13844 | -0.021 | -0.4226 | No | ||
| 133 | HOXC6 | 22339 2302 | 13875 | -0.021 | -0.4228 | No | ||
| 134 | SOX3 | 24145 | 13906 | -0.021 | -0.4230 | No | ||
| 135 | RNASE4 | 12218 2843 | 14145 | -0.024 | -0.4343 | No | ||
| 136 | SLC5A7 | 22938 | 14199 | -0.025 | -0.4355 | No | ||
| 137 | FGFR4 | 3226 | 14283 | -0.026 | -0.4382 | No | ||
| 138 | HESX1 | 22070 | 14666 | -0.033 | -0.4566 | No | ||
| 139 | HTR7 | 23690 | 14690 | -0.034 | -0.4556 | No | ||
| 140 | LGALS2 | 22213 | 14899 | -0.039 | -0.4643 | No | ||
| 141 | G6PC | 20656 | 15012 | -0.041 | -0.4676 | No | ||
| 142 | ISL1 | 21338 | 15026 | -0.042 | -0.4655 | No | ||
| 143 | FXYD2 | 3038 19470 3068 3039 | 15058 | -0.042 | -0.4643 | No | ||
| 144 | HOXC4 | 4864 | 15189 | -0.046 | -0.4683 | No | ||
| 145 | ZNF148 | 5949 | 15258 | -0.048 | -0.4687 | No | ||
| 146 | CS | 19839 | 15329 | -0.050 | -0.4691 | No | ||
| 147 | TCEA3 | 16031 | 15680 | -0.064 | -0.4838 | No | ||
| 148 | WBP4 | 21741 | 15969 | -0.078 | -0.4941 | No | ||
| 149 | SFRS7 | 22889 | 16140 | -0.088 | -0.4974 | No | ||
| 150 | KIF12 | 15865 | 16215 | -0.093 | -0.4952 | No | ||
| 151 | NR2F1 | 7015 21401 | 16363 | -0.103 | -0.4962 | No | ||
| 152 | RAB3IP | 19623 | 16408 | -0.106 | -0.4915 | No | ||
| 153 | BCL9 | 15232 | 16512 | -0.114 | -0.4894 | No | ||
| 154 | USP3 | 19074 | 17361 | -0.205 | -0.5216 | Yes | ||
| 155 | MSH5 | 23010 1599 | 17444 | -0.217 | -0.5115 | Yes | ||
| 156 | ZFPM2 | 22483 | 17829 | -0.281 | -0.5134 | Yes | ||
| 157 | CHCHD7 | 16280 | 17881 | -0.294 | -0.4964 | Yes | ||
| 158 | ARL6IP2 | 1496 7090 12093 | 17895 | -0.297 | -0.4771 | Yes | ||
| 159 | EGR1 | 23598 | 18044 | -0.337 | -0.4625 | Yes | ||
| 160 | BMI1 | 4448 15113 | 18088 | -0.351 | -0.4413 | Yes | ||
| 161 | TLE4 | 23720 3697 | 18457 | -0.763 | -0.4099 | Yes | ||
| 162 | E2F6 | 6925 6926 11920 | 18498 | -0.956 | -0.3478 | Yes | ||
| 163 | PABPC1 | 5219 9522 9523 23572 | 18615 | -5.269 | 0.0001 | Yes |