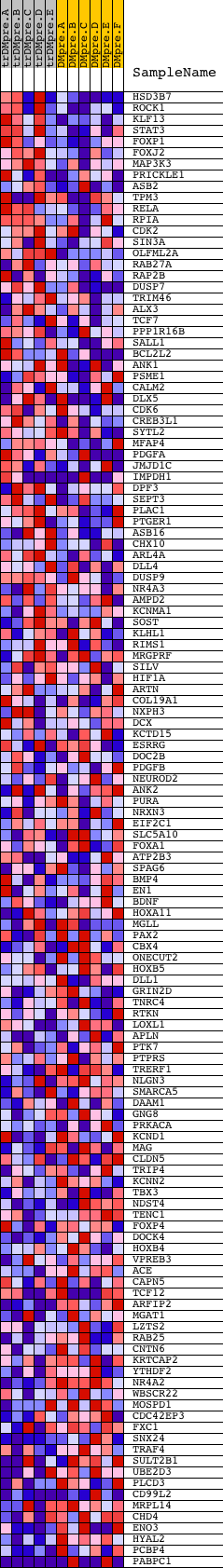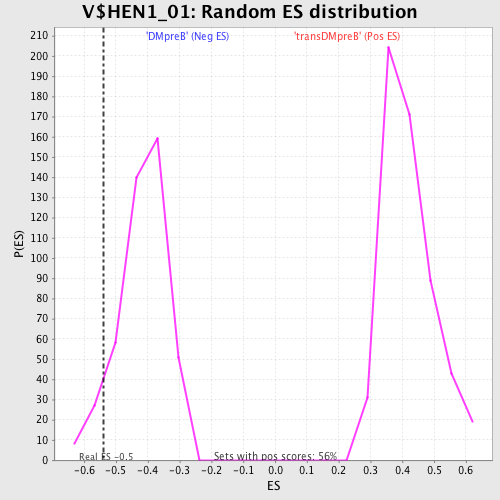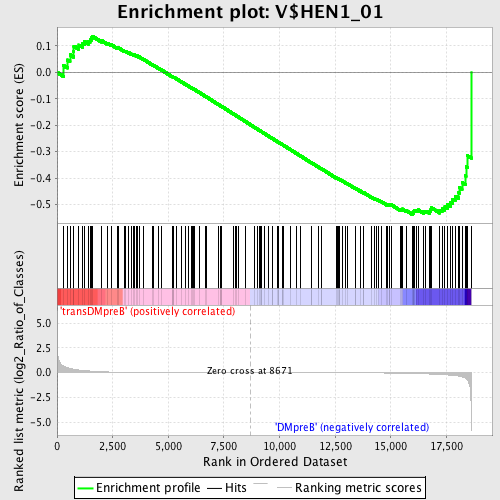
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$HEN1_01 |
| Enrichment Score (ES) | -0.5377753 |
| Normalized Enrichment Score (NES) | -1.2925767 |
| Nominal p-value | 0.07674944 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HSD3B7 | 4158 | 279 | 0.666 | 0.0259 | No | ||
| 2 | ROCK1 | 5386 | 458 | 0.495 | 0.0468 | No | ||
| 3 | KLF13 | 17807 | 580 | 0.428 | 0.0666 | No | ||
| 4 | STAT3 | 5525 9906 | 746 | 0.342 | 0.0787 | No | ||
| 5 | FOXP1 | 4242 | 755 | 0.338 | 0.0991 | No | ||
| 6 | FOXJ2 | 17294 | 981 | 0.268 | 0.1034 | No | ||
| 7 | MAP3K3 | 20626 | 1150 | 0.222 | 0.1080 | No | ||
| 8 | PRICKLE1 | 8379 | 1226 | 0.206 | 0.1166 | No | ||
| 9 | ASB2 | 20996 | 1409 | 0.173 | 0.1175 | No | ||
| 10 | TPM3 | 12233 7209 7208 1790 | 1480 | 0.161 | 0.1236 | No | ||
| 11 | RELA | 23783 | 1532 | 0.154 | 0.1304 | No | ||
| 12 | RPIA | 17124 | 1578 | 0.148 | 0.1370 | No | ||
| 13 | CDK2 | 3438 3373 19592 3322 | 1996 | 0.093 | 0.1202 | No | ||
| 14 | SIN3A | 5442 | 2261 | 0.070 | 0.1102 | No | ||
| 15 | OLFML2A | 6304 | 2438 | 0.060 | 0.1044 | No | ||
| 16 | RAB27A | 19381 | 2716 | 0.046 | 0.0922 | No | ||
| 17 | RAP2B | 7881 15580 | 2739 | 0.045 | 0.0938 | No | ||
| 18 | DUSP7 | 10661 | 3031 | 0.036 | 0.0803 | No | ||
| 19 | TRIM46 | 1779 15281 | 3074 | 0.035 | 0.0802 | No | ||
| 20 | ALX3 | 15456 | 3221 | 0.031 | 0.0742 | No | ||
| 21 | TCF7 | 1467 20466 | 3342 | 0.028 | 0.0694 | No | ||
| 22 | PPP1R16B | 6014 | 3428 | 0.026 | 0.0664 | No | ||
| 23 | SALL1 | 18796 12207 | 3456 | 0.026 | 0.0666 | No | ||
| 24 | BCL2L2 | 8653 4441 | 3589 | 0.024 | 0.0609 | No | ||
| 25 | ANK1 | 22501 18650 4385 8590 18649 | 3591 | 0.024 | 0.0623 | No | ||
| 26 | PSME1 | 5300 9637 | 3680 | 0.022 | 0.0589 | No | ||
| 27 | CALM2 | 8681 | 3889 | 0.019 | 0.0488 | No | ||
| 28 | DLX5 | 17219 1058 | 4277 | 0.015 | 0.0288 | No | ||
| 29 | CDK6 | 16600 | 4339 | 0.015 | 0.0264 | No | ||
| 30 | CREB3L1 | 14521 | 4563 | 0.013 | 0.0151 | No | ||
| 31 | SYTL2 | 18190 1539 3730 | 4675 | 0.012 | 0.0099 | No | ||
| 32 | MFAP4 | 20853 | 5169 | 0.009 | -0.0162 | No | ||
| 33 | PDGFA | 5237 | 5197 | 0.009 | -0.0171 | No | ||
| 34 | JMJD1C | 11531 19996 | 5228 | 0.009 | -0.0181 | No | ||
| 35 | IMPDH1 | 17197 1131 | 5232 | 0.009 | -0.0177 | No | ||
| 36 | DPF3 | 7659 12853 | 5360 | 0.009 | -0.0241 | No | ||
| 37 | SEPT3 | 6275 10775 | 5610 | 0.008 | -0.0371 | No | ||
| 38 | PLAC1 | 12079 | 5749 | 0.007 | -0.0441 | No | ||
| 39 | PTGER1 | 5309 | 5920 | 0.007 | -0.0529 | No | ||
| 40 | ASB16 | 20644 | 6041 | 0.006 | -0.0590 | No | ||
| 41 | CHX10 | 21208 | 6074 | 0.006 | -0.0604 | No | ||
| 42 | ARL4A | 8627 | 6114 | 0.006 | -0.0621 | No | ||
| 43 | DLL4 | 7040 | 6155 | 0.006 | -0.0639 | No | ||
| 44 | DUSP9 | 24307 | 6396 | 0.005 | -0.0766 | No | ||
| 45 | NR4A3 | 9473 16212 5183 | 6652 | 0.004 | -0.0901 | No | ||
| 46 | AMPD2 | 1931 15197 | 6696 | 0.004 | -0.0922 | No | ||
| 47 | KCNMA1 | 4947 | 7243 | 0.003 | -0.1215 | No | ||
| 48 | SOST | 20210 | 7342 | 0.003 | -0.1266 | No | ||
| 49 | KLHL1 | 21735 | 7358 | 0.003 | -0.1273 | No | ||
| 50 | RIMS1 | 8575 | 7371 | 0.003 | -0.1278 | No | ||
| 51 | MRGPRF | 17995 | 7389 | 0.003 | -0.1285 | No | ||
| 52 | SILV | 16130 | 7941 | 0.001 | -0.1582 | No | ||
| 53 | HIF1A | 4850 | 8017 | 0.001 | -0.1622 | No | ||
| 54 | ARTN | 15783 2430 2532 | 8070 | 0.001 | -0.1650 | No | ||
| 55 | COL19A1 | 4541 | 8174 | 0.001 | -0.1705 | No | ||
| 56 | NXPH3 | 20281 | 8463 | 0.000 | -0.1860 | No | ||
| 57 | DCX | 24040 8841 4604 | 8883 | -0.000 | -0.2087 | No | ||
| 58 | KCTD15 | 17864 | 9012 | -0.001 | -0.2156 | No | ||
| 59 | ESRRG | 6447 | 9087 | -0.001 | -0.2195 | No | ||
| 60 | DOC2B | 20347 8859 | 9155 | -0.001 | -0.2231 | No | ||
| 61 | PDGFB | 22199 2311 | 9170 | -0.001 | -0.2238 | No | ||
| 62 | NEUROD2 | 20262 | 9202 | -0.001 | -0.2254 | No | ||
| 63 | ANK2 | 4275 | 9300 | -0.001 | -0.2305 | No | ||
| 64 | PURA | 9670 | 9497 | -0.002 | -0.2410 | No | ||
| 65 | NRXN3 | 2153 5196 | 9670 | -0.002 | -0.2502 | No | ||
| 66 | EIF2C1 | 10672 | 9701 | -0.002 | -0.2517 | No | ||
| 67 | SLC5A10 | 20412 | 9889 | -0.003 | -0.2616 | No | ||
| 68 | FOXA1 | 21060 | 9936 | -0.003 | -0.2639 | No | ||
| 69 | ATP2B3 | 24308 11557 | 9965 | -0.003 | -0.2653 | No | ||
| 70 | SPAG6 | 22650 | 10114 | -0.003 | -0.2731 | No | ||
| 71 | BMP4 | 21863 | 10148 | -0.003 | -0.2747 | No | ||
| 72 | EN1 | 387 14155 | 10175 | -0.003 | -0.2759 | No | ||
| 73 | BDNF | 14926 2797 | 10471 | -0.004 | -0.2916 | No | ||
| 74 | HOXA11 | 17146 | 10774 | -0.005 | -0.3076 | No | ||
| 75 | MGLL | 17368 1145 | 10928 | -0.005 | -0.3156 | No | ||
| 76 | PAX2 | 9530 24416 | 11416 | -0.007 | -0.3415 | No | ||
| 77 | CBX4 | 20128 | 11433 | -0.007 | -0.3420 | No | ||
| 78 | ONECUT2 | 10346 | 11762 | -0.008 | -0.3592 | No | ||
| 79 | HOXB5 | 20687 | 11882 | -0.008 | -0.3652 | No | ||
| 80 | DLL1 | 23119 | 11884 | -0.008 | -0.3647 | No | ||
| 81 | GRIN2D | 9042 | 12538 | -0.011 | -0.3994 | No | ||
| 82 | TNRC4 | 15514 | 12597 | -0.011 | -0.4018 | No | ||
| 83 | RTKN | 17394 1156 | 12629 | -0.012 | -0.4028 | No | ||
| 84 | LOXL1 | 5002 | 12690 | -0.012 | -0.4053 | No | ||
| 85 | APLN | 24164 | 12694 | -0.012 | -0.4047 | No | ||
| 86 | PTK7 | 22959 | 12839 | -0.013 | -0.4117 | No | ||
| 87 | PTPRS | 1507 1565 1602 22928 | 12984 | -0.014 | -0.4187 | No | ||
| 88 | TRERF1 | 1546 23213 | 13044 | -0.014 | -0.4210 | No | ||
| 89 | NLGN3 | 10878 | 13401 | -0.017 | -0.4392 | No | ||
| 90 | SMARCA5 | 8215 | 13421 | -0.017 | -0.4392 | No | ||
| 91 | DAAM1 | 5536 | 13616 | -0.019 | -0.4486 | No | ||
| 92 | GNG8 | 18378 | 13775 | -0.020 | -0.4559 | No | ||
| 93 | PRKACA | 18549 3844 | 13785 | -0.020 | -0.4551 | No | ||
| 94 | KCND1 | 4940 | 14146 | -0.024 | -0.4731 | No | ||
| 95 | MAG | 17880 | 14274 | -0.026 | -0.4784 | No | ||
| 96 | CLDN5 | 22826 | 14356 | -0.028 | -0.4811 | No | ||
| 97 | TRIP4 | 12123 | 14450 | -0.029 | -0.4843 | No | ||
| 98 | KCNN2 | 8933 1949 | 14570 | -0.031 | -0.4888 | No | ||
| 99 | TBX3 | 16723 | 14823 | -0.037 | -0.5001 | No | ||
| 100 | NDST4 | 12259 7228 | 14841 | -0.037 | -0.4988 | No | ||
| 101 | TENC1 | 22349 9927 | 14944 | -0.040 | -0.5018 | No | ||
| 102 | FOXP4 | 22949 | 14958 | -0.040 | -0.5001 | No | ||
| 103 | DOCK4 | 10705 | 15019 | -0.041 | -0.5007 | No | ||
| 104 | HOXB4 | 20686 | 15426 | -0.053 | -0.5194 | No | ||
| 105 | VPREB3 | 19984 | 15491 | -0.056 | -0.5194 | No | ||
| 106 | ACE | 1419 4313 | 15510 | -0.057 | -0.5169 | No | ||
| 107 | CAPN5 | 17752 | 15697 | -0.065 | -0.5230 | No | ||
| 108 | TCF12 | 10044 3125 | 15972 | -0.078 | -0.5330 | Yes | ||
| 109 | ARFIP2 | 1226 17702 | 16025 | -0.081 | -0.5308 | Yes | ||
| 110 | MGAT1 | 9384 | 16033 | -0.081 | -0.5261 | Yes | ||
| 111 | LZTS2 | 10362 | 16068 | -0.083 | -0.5228 | Yes | ||
| 112 | RAB25 | 15287 | 16138 | -0.088 | -0.5212 | Yes | ||
| 113 | CNTN6 | 17346 | 16227 | -0.094 | -0.5202 | Yes | ||
| 114 | KRTCAP2 | 15541 | 16470 | -0.111 | -0.5264 | Yes | ||
| 115 | YTHDF2 | 5623 5624 | 16574 | -0.119 | -0.5246 | Yes | ||
| 116 | NR4A2 | 5203 | 16755 | -0.136 | -0.5260 | Yes | ||
| 117 | WBSCR22 | 3602 3561 16347 | 16799 | -0.142 | -0.5196 | Yes | ||
| 118 | MOSPD1 | 12868 12869 7672 | 16807 | -0.143 | -0.5111 | Yes | ||
| 119 | CDC42EP3 | 6436 | 17200 | -0.185 | -0.5209 | Yes | ||
| 120 | FXC1 | 8988 4043 | 17305 | -0.198 | -0.5144 | Yes | ||
| 121 | SNX24 | 23553 | 17433 | -0.214 | -0.5080 | Yes | ||
| 122 | TRAF4 | 10217 5796 1400 | 17558 | -0.232 | -0.5005 | Yes | ||
| 123 | SULT2B1 | 2016 17827 | 17687 | -0.251 | -0.4919 | Yes | ||
| 124 | UBE2D3 | 7253 | 17768 | -0.268 | -0.4797 | Yes | ||
| 125 | PLCD3 | 20200 | 17892 | -0.297 | -0.4681 | Yes | ||
| 126 | CD99L2 | 2612 24141 | 18054 | -0.340 | -0.4559 | Yes | ||
| 127 | MRPL14 | 23223 | 18074 | -0.347 | -0.4356 | Yes | ||
| 128 | CHD4 | 8418 17281 4225 | 18201 | -0.422 | -0.4164 | Yes | ||
| 129 | ENO3 | 8905 | 18359 | -0.547 | -0.3912 | Yes | ||
| 130 | HYAL2 | 4891 | 18381 | -0.581 | -0.3566 | Yes | ||
| 131 | PCBP4 | 12235 | 18443 | -0.725 | -0.3152 | Yes | ||
| 132 | PABPC1 | 5219 9522 9523 23572 | 18615 | -5.269 | 0.0001 | Yes |