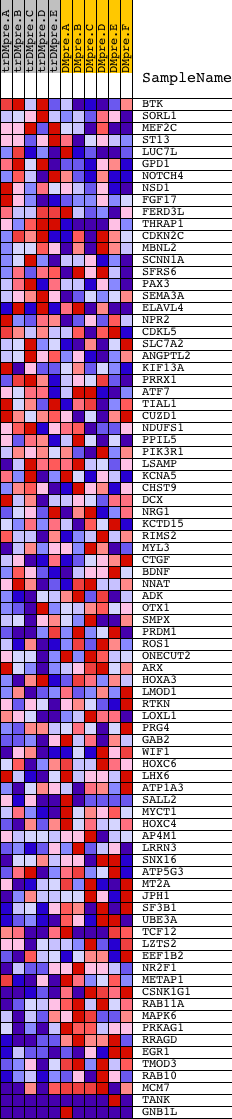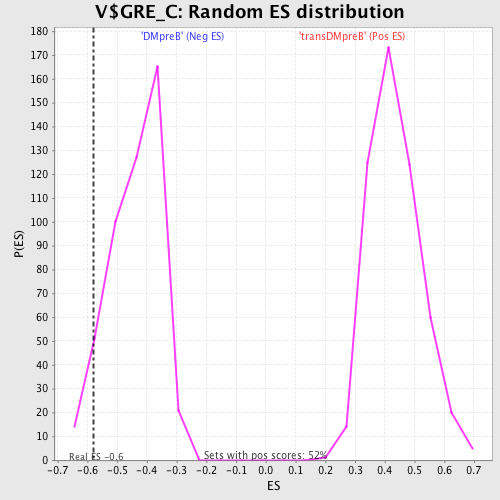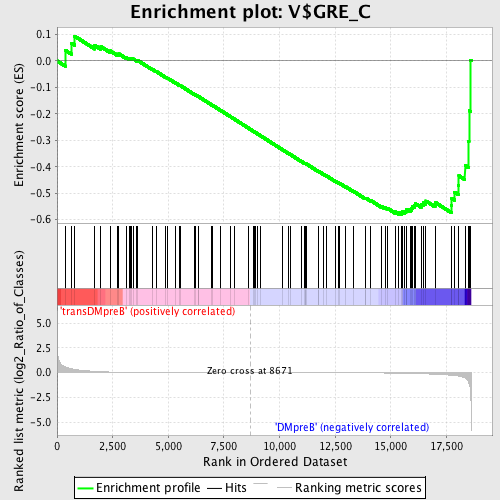
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$GRE_C |
| Enrichment Score (ES) | -0.5803954 |
| Normalized Enrichment Score (NES) | -1.3196822 |
| Nominal p-value | 0.06903765 |
| FDR q-value | 0.98899734 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BTK | 24061 | 369 | 0.566 | 0.0401 | No | ||
| 2 | SORL1 | 5474 | 639 | 0.391 | 0.0670 | No | ||
| 3 | MEF2C | 3204 9378 | 787 | 0.327 | 0.0937 | No | ||
| 4 | ST13 | 12865 | 1693 | 0.130 | 0.0586 | No | ||
| 5 | LUC7L | 47 1520 | 1969 | 0.096 | 0.0539 | No | ||
| 6 | GPD1 | 22361 | 2391 | 0.063 | 0.0378 | No | ||
| 7 | NOTCH4 | 23277 9475 | 2734 | 0.045 | 0.0242 | No | ||
| 8 | NSD1 | 2134 5197 | 2777 | 0.044 | 0.0266 | No | ||
| 9 | FGF17 | 21757 | 3120 | 0.034 | 0.0117 | No | ||
| 10 | FERD3L | 21292 | 3235 | 0.031 | 0.0088 | No | ||
| 11 | THRAP1 | 20309 | 3304 | 0.029 | 0.0082 | No | ||
| 12 | CDKN2C | 15806 2321 | 3336 | 0.028 | 0.0095 | No | ||
| 13 | MBNL2 | 21932 | 3427 | 0.026 | 0.0074 | No | ||
| 14 | SCNN1A | 17278 | 3582 | 0.024 | 0.0016 | No | ||
| 15 | SFRS6 | 14751 | 3612 | 0.023 | 0.0025 | No | ||
| 16 | PAX3 | 13910 | 4275 | 0.015 | -0.0316 | No | ||
| 17 | SEMA3A | 16919 | 4448 | 0.014 | -0.0395 | No | ||
| 18 | ELAVL4 | 15805 4889 9137 | 4871 | 0.011 | -0.0611 | No | ||
| 19 | NPR2 | 16230 | 4966 | 0.011 | -0.0650 | No | ||
| 20 | CDKL5 | 24019 | 5340 | 0.009 | -0.0842 | No | ||
| 21 | SLC7A2 | 4431 8644 | 5488 | 0.008 | -0.0913 | No | ||
| 22 | ANGPTL2 | 15032 18104 | 5529 | 0.008 | -0.0926 | No | ||
| 23 | KIF13A | 21475 | 6166 | 0.006 | -0.1263 | No | ||
| 24 | PRRX1 | 9595 5271 | 6205 | 0.006 | -0.1277 | No | ||
| 25 | ATF7 | 10303 5869 | 6352 | 0.005 | -0.1351 | No | ||
| 26 | TIAL1 | 996 5753 | 6368 | 0.005 | -0.1353 | No | ||
| 27 | CUZD1 | 17598 | 6952 | 0.004 | -0.1664 | No | ||
| 28 | NDUFS1 | 13940 | 6973 | 0.004 | -0.1671 | No | ||
| 29 | PPIL5 | 21255 | 7336 | 0.003 | -0.1863 | No | ||
| 30 | PIK3R1 | 3170 | 7781 | 0.002 | -0.2101 | No | ||
| 31 | LSAMP | 1690 6500 | 7989 | 0.001 | -0.2211 | No | ||
| 32 | KCNA5 | 9203 | 8590 | 0.000 | -0.2535 | No | ||
| 33 | CHST9 | 7727 | 8831 | -0.000 | -0.2664 | No | ||
| 34 | DCX | 24040 8841 4604 | 8883 | -0.000 | -0.2691 | No | ||
| 35 | NRG1 | 3885 | 8910 | -0.000 | -0.2705 | No | ||
| 36 | KCTD15 | 17864 | 9012 | -0.001 | -0.2758 | No | ||
| 37 | RIMS2 | 4370 | 9139 | -0.001 | -0.2825 | No | ||
| 38 | MYL3 | 19291 | 10126 | -0.003 | -0.3354 | No | ||
| 39 | CTGF | 20068 | 10412 | -0.004 | -0.3504 | No | ||
| 40 | BDNF | 14926 2797 | 10471 | -0.004 | -0.3531 | No | ||
| 41 | NNAT | 2764 5180 | 10991 | -0.005 | -0.3805 | No | ||
| 42 | ADK | 3302 3454 8555 | 11116 | -0.006 | -0.3866 | No | ||
| 43 | OTX1 | 20517 | 11148 | -0.006 | -0.3876 | No | ||
| 44 | SMPX | 24220 | 11161 | -0.006 | -0.3877 | No | ||
| 45 | PRDM1 | 19775 3337 | 11218 | -0.006 | -0.3901 | No | ||
| 46 | ROS1 | 19771 | 11751 | -0.008 | -0.4179 | No | ||
| 47 | ONECUT2 | 10346 | 11762 | -0.008 | -0.4177 | No | ||
| 48 | ARX | 24290 | 11970 | -0.008 | -0.4279 | No | ||
| 49 | HOXA3 | 9108 1075 | 12121 | -0.009 | -0.4351 | No | ||
| 50 | LMOD1 | 14119 | 12516 | -0.011 | -0.4552 | No | ||
| 51 | RTKN | 17394 1156 | 12629 | -0.012 | -0.4600 | No | ||
| 52 | LOXL1 | 5002 | 12690 | -0.012 | -0.4620 | No | ||
| 53 | PRG4 | 13592 4036 | 12982 | -0.014 | -0.4762 | No | ||
| 54 | GAB2 | 1821 18184 2025 | 13305 | -0.016 | -0.4919 | No | ||
| 55 | WIF1 | 19866 | 13853 | -0.021 | -0.5192 | No | ||
| 56 | HOXC6 | 22339 2302 | 13875 | -0.021 | -0.5181 | No | ||
| 57 | LHX6 | 9275 2914 | 14105 | -0.024 | -0.5279 | No | ||
| 58 | ATP1A3 | 21 | 14577 | -0.032 | -0.5500 | No | ||
| 59 | SALL2 | 21840 | 14743 | -0.035 | -0.5552 | No | ||
| 60 | MYCT1 | 7572 | 14833 | -0.037 | -0.5561 | No | ||
| 61 | HOXC4 | 4864 | 15189 | -0.046 | -0.5704 | No | ||
| 62 | AP4M1 | 3499 16656 | 15358 | -0.051 | -0.5740 | No | ||
| 63 | LRRN3 | 21077 | 15477 | -0.055 | -0.5745 | Yes | ||
| 64 | SNX16 | 15374 | 15514 | -0.057 | -0.5704 | Yes | ||
| 65 | ATP5G3 | 14558 | 15609 | -0.061 | -0.5690 | Yes | ||
| 66 | MT2A | 18523 | 15683 | -0.064 | -0.5661 | Yes | ||
| 67 | JPH1 | 13994 | 15713 | -0.066 | -0.5607 | Yes | ||
| 68 | SF3B1 | 13954 | 15878 | -0.074 | -0.5618 | Yes | ||
| 69 | UBE3A | 1209 1084 1830 | 15942 | -0.077 | -0.5570 | Yes | ||
| 70 | TCF12 | 10044 3125 | 15972 | -0.078 | -0.5503 | Yes | ||
| 71 | LZTS2 | 10362 | 16068 | -0.083 | -0.5466 | Yes | ||
| 72 | EEF1B2 | 4131 12063 | 16101 | -0.085 | -0.5392 | Yes | ||
| 73 | NR2F1 | 7015 21401 | 16363 | -0.103 | -0.5425 | Yes | ||
| 74 | METAP1 | 15154 | 16460 | -0.111 | -0.5359 | Yes | ||
| 75 | CSNK1G1 | 3057 5671 19397 | 16576 | -0.120 | -0.5294 | Yes | ||
| 76 | RAB11A | 7013 | 16990 | -0.164 | -0.5344 | Yes | ||
| 77 | MAPK6 | 19062 | 17726 | -0.259 | -0.5466 | Yes | ||
| 78 | PRKAG1 | 22135 | 17739 | -0.261 | -0.5196 | Yes | ||
| 79 | RRAGD | 11964 | 17865 | -0.291 | -0.4956 | Yes | ||
| 80 | EGR1 | 23598 | 18044 | -0.337 | -0.4695 | Yes | ||
| 81 | TMOD3 | 6945 | 18060 | -0.342 | -0.4341 | Yes | ||
| 82 | RAB10 | 2090 21119 | 18334 | -0.514 | -0.3945 | Yes | ||
| 83 | MCM7 | 9372 3568 | 18494 | -0.936 | -0.3039 | Yes | ||
| 84 | TANK | 2839 15006 2729 | 18515 | -1.112 | -0.1872 | Yes | ||
| 85 | GNB1L | 8921 22827 | 18576 | -1.818 | 0.0022 | Yes |