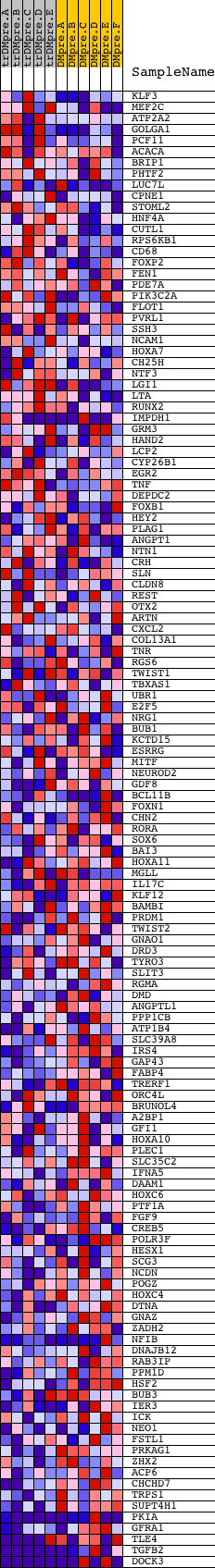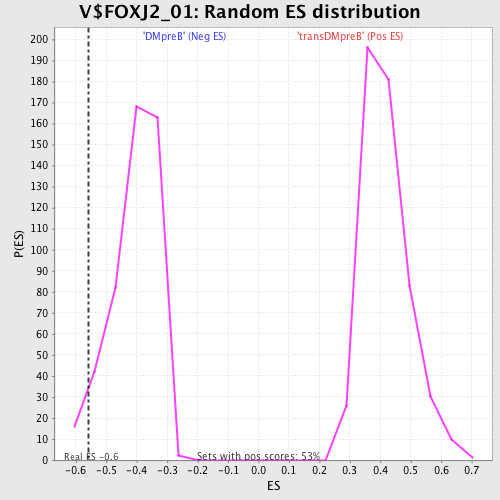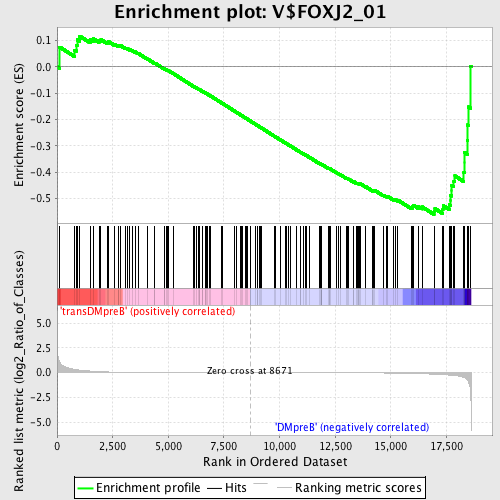
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$FOXJ2_01 |
| Enrichment Score (ES) | -0.5598524 |
| Normalized Enrichment Score (NES) | -1.375308 |
| Nominal p-value | 0.040169135 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KLF3 | 4961 | 117 | 1.062 | 0.0748 | No | ||
| 2 | MEF2C | 3204 9378 | 787 | 0.327 | 0.0635 | No | ||
| 3 | ATP2A2 | 4421 3481 | 863 | 0.303 | 0.0826 | No | ||
| 4 | GOLGA1 | 14595 | 904 | 0.292 | 0.1027 | No | ||
| 5 | PCF11 | 121 | 1016 | 0.256 | 0.1163 | No | ||
| 6 | ACACA | 309 4209 | 1489 | 0.160 | 0.1030 | No | ||
| 7 | BRIP1 | 20311 | 1625 | 0.140 | 0.1064 | No | ||
| 8 | PHTF2 | 16599 | 1886 | 0.106 | 0.1004 | No | ||
| 9 | LUC7L | 47 1520 | 1969 | 0.096 | 0.1032 | No | ||
| 10 | CPNE1 | 11186 | 2252 | 0.071 | 0.0934 | No | ||
| 11 | STOML2 | 15902 | 2295 | 0.068 | 0.0963 | No | ||
| 12 | HNF4A | 14746 | 2557 | 0.053 | 0.0862 | No | ||
| 13 | CUTL1 | 3507 3480 8820 3609 4577 | 2749 | 0.045 | 0.0793 | No | ||
| 14 | RPS6KB1 | 7815 1207 13040 | 2778 | 0.044 | 0.0811 | No | ||
| 15 | CD68 | 4501 | 2830 | 0.042 | 0.0816 | No | ||
| 16 | FOXP2 | 4312 17523 8523 | 3091 | 0.034 | 0.0701 | No | ||
| 17 | FEN1 | 8961 | 3149 | 0.033 | 0.0696 | No | ||
| 18 | PDE7A | 9544 1788 | 3270 | 0.030 | 0.0654 | No | ||
| 19 | PIK3C2A | 17667 | 3377 | 0.027 | 0.0617 | No | ||
| 20 | FLOT1 | 23249 1613 | 3507 | 0.025 | 0.0566 | No | ||
| 21 | PVRL1 | 3006 19482 12211 7192 | 3636 | 0.023 | 0.0514 | No | ||
| 22 | SSH3 | 3709 10890 | 4063 | 0.017 | 0.0297 | No | ||
| 23 | NCAM1 | 5149 | 4368 | 0.014 | 0.0144 | No | ||
| 24 | HOXA7 | 4862 | 4398 | 0.014 | 0.0139 | No | ||
| 25 | CH25H | 23695 | 4827 | 0.011 | -0.0084 | No | ||
| 26 | NTF3 | 16991 | 4830 | 0.011 | -0.0077 | No | ||
| 27 | LGI1 | 23867 | 4903 | 0.011 | -0.0107 | No | ||
| 28 | LTA | 23003 | 4961 | 0.011 | -0.0130 | No | ||
| 29 | RUNX2 | 4480 8700 | 5017 | 0.010 | -0.0152 | No | ||
| 30 | IMPDH1 | 17197 1131 | 5232 | 0.009 | -0.0261 | No | ||
| 31 | GRM3 | 16929 | 6126 | 0.006 | -0.0740 | No | ||
| 32 | HAND2 | 18615 | 6162 | 0.006 | -0.0754 | No | ||
| 33 | LCP2 | 4988 9268 | 6272 | 0.005 | -0.0809 | No | ||
| 34 | CYP26B1 | 17093 | 6357 | 0.005 | -0.0850 | No | ||
| 35 | EGR2 | 8886 | 6388 | 0.005 | -0.0862 | No | ||
| 36 | TNF | 23004 | 6407 | 0.005 | -0.0868 | No | ||
| 37 | DEPDC2 | 14295 | 6550 | 0.005 | -0.0942 | No | ||
| 38 | FOXB1 | 12254 19069 | 6554 | 0.005 | -0.0940 | No | ||
| 39 | HEY2 | 19794 | 6654 | 0.004 | -0.0990 | No | ||
| 40 | PLAG1 | 7148 15949 12161 | 6690 | 0.004 | -0.1005 | No | ||
| 41 | ANGPT1 | 8588 2260 22303 | 6701 | 0.004 | -0.1008 | No | ||
| 42 | NTN1 | 20400 | 6762 | 0.004 | -0.1037 | No | ||
| 43 | CRH | 8784 | 6828 | 0.004 | -0.1069 | No | ||
| 44 | SLN | 19450 | 6880 | 0.004 | -0.1094 | No | ||
| 45 | CLDN8 | 22549 1747 | 7388 | 0.003 | -0.1366 | No | ||
| 46 | REST | 16818 | 7431 | 0.003 | -0.1387 | No | ||
| 47 | OTX2 | 9519 | 7990 | 0.001 | -0.1688 | No | ||
| 48 | ARTN | 15783 2430 2532 | 8070 | 0.001 | -0.1729 | No | ||
| 49 | CXCL2 | 9790 | 8251 | 0.001 | -0.1826 | No | ||
| 50 | COL13A1 | 8763 | 8288 | 0.001 | -0.1845 | No | ||
| 51 | TNR | 5787 | 8354 | 0.001 | -0.1880 | No | ||
| 52 | RGS6 | 21219 | 8457 | 0.000 | -0.1935 | No | ||
| 53 | TWIST1 | 21291 | 8501 | 0.000 | -0.1957 | No | ||
| 54 | TBXAS1 | 17480 1105 | 8565 | 0.000 | -0.1991 | No | ||
| 55 | UBR1 | 5828 | 8690 | -0.000 | -0.2058 | No | ||
| 56 | E2F5 | 15633 | 8699 | -0.000 | -0.2063 | No | ||
| 57 | NRG1 | 3885 | 8910 | -0.000 | -0.2176 | No | ||
| 58 | BUB1 | 8665 | 9007 | -0.001 | -0.2227 | No | ||
| 59 | KCTD15 | 17864 | 9012 | -0.001 | -0.2229 | No | ||
| 60 | ESRRG | 6447 | 9087 | -0.001 | -0.2268 | No | ||
| 61 | MITF | 17349 | 9127 | -0.001 | -0.2289 | No | ||
| 62 | NEUROD2 | 20262 | 9202 | -0.001 | -0.2328 | No | ||
| 63 | GDF8 | 9411 | 9791 | -0.002 | -0.2644 | No | ||
| 64 | BCL11B | 7190 | 9826 | -0.003 | -0.2661 | No | ||
| 65 | FOXN1 | 20337 | 10052 | -0.003 | -0.2780 | No | ||
| 66 | CHN2 | 7652 | 10284 | -0.004 | -0.2902 | No | ||
| 67 | RORA | 3019 5388 | 10302 | -0.004 | -0.2909 | No | ||
| 68 | SOX6 | 3684 5481 9851 | 10391 | -0.004 | -0.2953 | No | ||
| 69 | BAI3 | 5580 | 10501 | -0.004 | -0.3009 | No | ||
| 70 | HOXA11 | 17146 | 10774 | -0.005 | -0.3153 | No | ||
| 71 | MGLL | 17368 1145 | 10928 | -0.005 | -0.3231 | No | ||
| 72 | IL17C | 18438 | 10946 | -0.005 | -0.3237 | No | ||
| 73 | KLF12 | 4960 2637 21732 9228 | 11065 | -0.006 | -0.3296 | No | ||
| 74 | BAMBI | 23629 | 11183 | -0.006 | -0.3355 | No | ||
| 75 | PRDM1 | 19775 3337 | 11218 | -0.006 | -0.3369 | No | ||
| 76 | TWIST2 | 14185 | 11334 | -0.006 | -0.3426 | No | ||
| 77 | GNAO1 | 4785 3829 | 11779 | -0.008 | -0.3660 | No | ||
| 78 | DRD3 | 22750 | 11826 | -0.008 | -0.3679 | No | ||
| 79 | TYRO3 | 5811 | 11880 | -0.008 | -0.3702 | No | ||
| 80 | SLIT3 | 20925 | 11886 | -0.008 | -0.3698 | No | ||
| 81 | RGMA | 18213 | 11888 | -0.008 | -0.3693 | No | ||
| 82 | DMD | 24295 2647 | 12181 | -0.009 | -0.3843 | No | ||
| 83 | ANGPTL1 | 10195 8589 13064 | 12225 | -0.010 | -0.3859 | No | ||
| 84 | PPP1CB | 5282 3529 3504 | 12226 | -0.010 | -0.3852 | No | ||
| 85 | ATP1B4 | 12586 | 12270 | -0.010 | -0.3868 | No | ||
| 86 | SLC39A8 | 12547 1776 | 12547 | -0.011 | -0.4009 | No | ||
| 87 | IRS4 | 9183 4926 | 12643 | -0.012 | -0.4051 | No | ||
| 88 | GAP43 | 9001 | 12726 | -0.012 | -0.4086 | No | ||
| 89 | FABP4 | 8601 | 13011 | -0.014 | -0.4229 | No | ||
| 90 | TRERF1 | 1546 23213 | 13044 | -0.014 | -0.4236 | No | ||
| 91 | ORC4L | 11172 6460 | 13098 | -0.014 | -0.4254 | No | ||
| 92 | BRUNOL4 | 2010 4229 | 13325 | -0.016 | -0.4364 | No | ||
| 93 | A2BP1 | 11212 1652 1689 | 13329 | -0.016 | -0.4353 | No | ||
| 94 | GFI1 | 16451 | 13461 | -0.017 | -0.4411 | No | ||
| 95 | HOXA10 | 17145 989 | 13494 | -0.017 | -0.4415 | No | ||
| 96 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 13549 | -0.018 | -0.4430 | No | ||
| 97 | SLC35C2 | 10463 14339 | 13550 | -0.018 | -0.4417 | No | ||
| 98 | IFNA5 | 9151 | 13605 | -0.018 | -0.4432 | No | ||
| 99 | DAAM1 | 5536 | 13616 | -0.019 | -0.4423 | No | ||
| 100 | HOXC6 | 22339 2302 | 13875 | -0.021 | -0.4547 | No | ||
| 101 | PTF1A | 15110 | 14194 | -0.025 | -0.4700 | No | ||
| 102 | FGF9 | 8966 | 14196 | -0.025 | -0.4681 | No | ||
| 103 | CREB5 | 10551 | 14233 | -0.026 | -0.4681 | No | ||
| 104 | POLR3F | 2814 7674 | 14287 | -0.026 | -0.4690 | No | ||
| 105 | HESX1 | 22070 | 14666 | -0.033 | -0.4869 | No | ||
| 106 | SCG3 | 19061 | 14795 | -0.036 | -0.4910 | No | ||
| 107 | NCDN | 15756 2487 6471 | 14856 | -0.038 | -0.4914 | No | ||
| 108 | POGZ | 6031 1911 | 15126 | -0.044 | -0.5025 | No | ||
| 109 | HOXC4 | 4864 | 15189 | -0.046 | -0.5024 | No | ||
| 110 | DTNA | 8870 4647 23611 1991 | 15315 | -0.050 | -0.5053 | No | ||
| 111 | GNAZ | 71 | 15908 | -0.075 | -0.5316 | No | ||
| 112 | ZADH2 | 23501 | 15985 | -0.079 | -0.5297 | No | ||
| 113 | NFIB | 15855 | 15999 | -0.079 | -0.5243 | No | ||
| 114 | DNAJB12 | 7146 | 16248 | -0.096 | -0.5305 | No | ||
| 115 | RAB3IP | 19623 | 16408 | -0.106 | -0.5310 | No | ||
| 116 | PPM1D | 20721 | 16943 | -0.158 | -0.5478 | Yes | ||
| 117 | HSF2 | 9128 | 16982 | -0.163 | -0.5374 | Yes | ||
| 118 | BUB3 | 18045 | 17339 | -0.202 | -0.5413 | Yes | ||
| 119 | IER3 | 23248 | 17349 | -0.204 | -0.5262 | Yes | ||
| 120 | ICK | 7135 12146 | 17641 | -0.244 | -0.5233 | Yes | ||
| 121 | NEO1 | 5161 | 17686 | -0.251 | -0.5065 | Yes | ||
| 122 | FSTL1 | 22765 | 17690 | -0.252 | -0.4874 | Yes | ||
| 123 | PRKAG1 | 22135 | 17739 | -0.261 | -0.4701 | Yes | ||
| 124 | ZHX2 | 11842 | 17741 | -0.262 | -0.4502 | Yes | ||
| 125 | ACP6 | 12394 | 17832 | -0.282 | -0.4335 | Yes | ||
| 126 | CHCHD7 | 16280 | 17881 | -0.294 | -0.4136 | Yes | ||
| 127 | TRPS1 | 8195 | 18282 | -0.473 | -0.3992 | Yes | ||
| 128 | SUPT4H1 | 9938 | 18315 | -0.497 | -0.3630 | Yes | ||
| 129 | PKIA | 15639 | 18316 | -0.497 | -0.3250 | Yes | ||
| 130 | GFRA1 | 9015 | 18437 | -0.707 | -0.2776 | Yes | ||
| 131 | TLE4 | 23720 3697 | 18457 | -0.763 | -0.2204 | Yes | ||
| 132 | TGFB2 | 5743 | 18487 | -0.915 | -0.1521 | Yes | ||
| 133 | DOCK3 | 9920 5537 10751 | 18584 | -2.084 | 0.0017 | Yes |