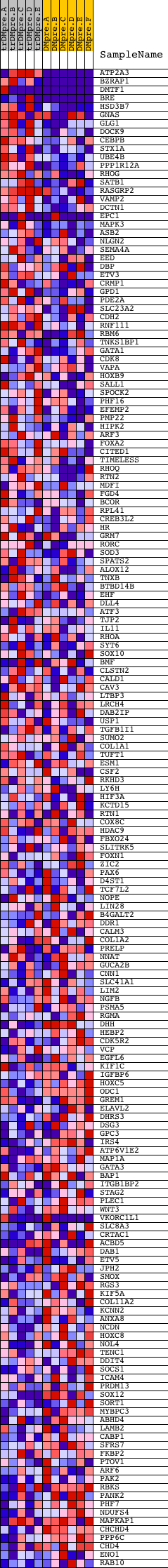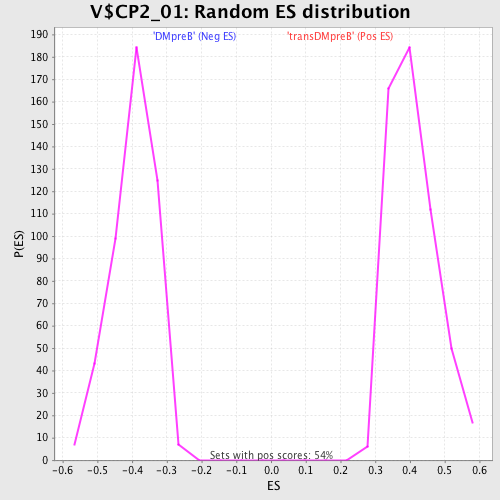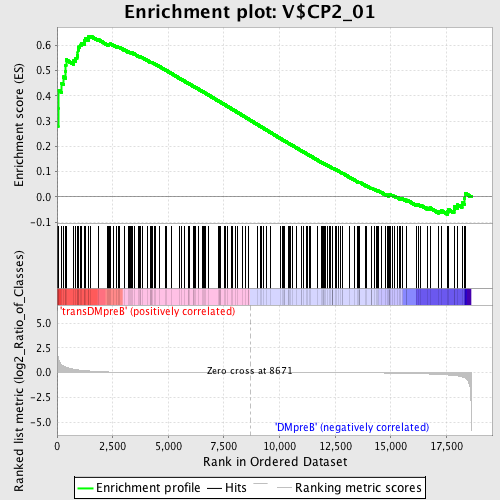
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$CP2_01 |
| Enrichment Score (ES) | 0.6366802 |
| Normalized Enrichment Score (NES) | 1.5677769 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.3577199 |
| FWER p-Value | 0.749 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 20795 1314 | 0 | 6.225 | 0.2815 | Yes | ||
| 2 | BZRAP1 | 9882 | 41 | 1.594 | 0.3514 | Yes | ||
| 3 | DMTF1 | 887 3662 16928 3530 | 46 | 1.574 | 0.4223 | Yes | ||
| 4 | BRE | 3640 4226 16879 | 209 | 0.780 | 0.4488 | Yes | ||
| 5 | HSD3B7 | 4158 | 279 | 0.666 | 0.4752 | Yes | ||
| 6 | GNAS | 9025 2963 2752 | 376 | 0.559 | 0.4952 | Yes | ||
| 7 | GLG1 | 5421 | 385 | 0.554 | 0.5199 | Yes | ||
| 8 | DOCK9 | 8369 | 401 | 0.537 | 0.5433 | Yes | ||
| 9 | CEBPB | 8733 | 751 | 0.341 | 0.5398 | Yes | ||
| 10 | STX1A | 16679 | 841 | 0.310 | 0.5490 | Yes | ||
| 11 | UBE4B | 2433 15672 | 894 | 0.294 | 0.5595 | Yes | ||
| 12 | PPP1R12A | 3354 19886 | 936 | 0.281 | 0.5700 | Yes | ||
| 13 | RHOG | 17727 | 938 | 0.280 | 0.5826 | Yes | ||
| 14 | SATB1 | 5408 | 961 | 0.272 | 0.5937 | Yes | ||
| 15 | RASGRP2 | 9693 | 1028 | 0.254 | 0.6016 | Yes | ||
| 16 | VAMP2 | 20826 | 1082 | 0.237 | 0.6094 | Yes | ||
| 17 | DCTN1 | 1100 17392 | 1228 | 0.206 | 0.6108 | Yes | ||
| 18 | EPC1 | 2039 8907 2033 | 1241 | 0.204 | 0.6194 | Yes | ||
| 19 | MAPK3 | 6458 11170 | 1268 | 0.198 | 0.6269 | Yes | ||
| 20 | ASB2 | 20996 | 1409 | 0.173 | 0.6272 | Yes | ||
| 21 | NLGN2 | 10117 | 1424 | 0.171 | 0.6341 | Yes | ||
| 22 | SEMA4A | 15289 | 1509 | 0.157 | 0.6367 | Yes | ||
| 23 | EED | 17759 | 1850 | 0.111 | 0.6233 | No | ||
| 24 | DBP | 18242 | 2274 | 0.069 | 0.6035 | No | ||
| 25 | ETV3 | 11294 | 2322 | 0.066 | 0.6039 | No | ||
| 26 | CRMP1 | 16860 | 2353 | 0.065 | 0.6052 | No | ||
| 27 | GPD1 | 22361 | 2391 | 0.063 | 0.6060 | No | ||
| 28 | PDE2A | 18170 | 2528 | 0.055 | 0.6011 | No | ||
| 29 | SLC23A2 | 14421 | 2681 | 0.047 | 0.5950 | No | ||
| 30 | CDH2 | 1963 8727 8726 4508 | 2780 | 0.044 | 0.5917 | No | ||
| 31 | RNF111 | 13555 8218 13556 | 2802 | 0.043 | 0.5925 | No | ||
| 32 | RBM6 | 9707 3095 3063 2995 | 3019 | 0.036 | 0.5824 | No | ||
| 33 | TNKS1BP1 | 14970 | 3189 | 0.032 | 0.5747 | No | ||
| 34 | GATA1 | 24196 | 3273 | 0.030 | 0.5715 | No | ||
| 35 | CDK8 | 6455 6456 3585 3542 | 3308 | 0.029 | 0.5710 | No | ||
| 36 | VAPA | 22908 | 3345 | 0.028 | 0.5703 | No | ||
| 37 | HOXB9 | 20689 | 3366 | 0.028 | 0.5704 | No | ||
| 38 | SALL1 | 18796 12207 | 3456 | 0.026 | 0.5668 | No | ||
| 39 | SPOCK2 | 13585 | 3657 | 0.023 | 0.5569 | No | ||
| 40 | PHF16 | 11795 6901 | 3715 | 0.022 | 0.5548 | No | ||
| 41 | EFEMP2 | 23781 | 3725 | 0.021 | 0.5553 | No | ||
| 42 | PMP22 | 9594 1312 | 3741 | 0.021 | 0.5555 | No | ||
| 43 | HIPK2 | 17180 | 3832 | 0.020 | 0.5515 | No | ||
| 44 | ARF3 | 22137 | 4073 | 0.017 | 0.5393 | No | ||
| 45 | FOXA2 | 14405 2889 | 4181 | 0.016 | 0.5342 | No | ||
| 46 | CITED1 | 24091 | 4212 | 0.016 | 0.5333 | No | ||
| 47 | TIMELESS | 10177 | 4231 | 0.016 | 0.5330 | No | ||
| 48 | RHOQ | 23142 | 4237 | 0.015 | 0.5334 | No | ||
| 49 | RTN2 | 9764 | 4290 | 0.015 | 0.5313 | No | ||
| 50 | MDFI | 22950 | 4360 | 0.014 | 0.5282 | No | ||
| 51 | FGD4 | 10306 5870 1717 1669 1700 | 4433 | 0.014 | 0.5249 | No | ||
| 52 | BCOR | 12934 2561 | 4585 | 0.013 | 0.5173 | No | ||
| 53 | RPL41 | 12611 | 4883 | 0.011 | 0.5017 | No | ||
| 54 | CREB3L2 | 17182 | 4934 | 0.011 | 0.4995 | No | ||
| 55 | HR | 3622 9119 | 5128 | 0.010 | 0.4894 | No | ||
| 56 | GRM7 | 17338 | 5514 | 0.008 | 0.4689 | No | ||
| 57 | RORC | 9732 | 5600 | 0.008 | 0.4647 | No | ||
| 58 | SOD3 | 16848 | 5706 | 0.007 | 0.4593 | No | ||
| 59 | SPATS2 | 13044 | 5884 | 0.007 | 0.4500 | No | ||
| 60 | ALOX12 | 20372 | 5900 | 0.007 | 0.4495 | No | ||
| 61 | TNXB | 1557 8174 878 8175 | 5971 | 0.006 | 0.4460 | No | ||
| 62 | BTBD14B | 7344 7343 | 6123 | 0.006 | 0.4380 | No | ||
| 63 | EHF | 4657 | 6135 | 0.006 | 0.4377 | No | ||
| 64 | DLL4 | 7040 | 6155 | 0.006 | 0.4370 | No | ||
| 65 | ATF3 | 13720 | 6232 | 0.006 | 0.4331 | No | ||
| 66 | TJP2 | 5761 3699 | 6336 | 0.005 | 0.4277 | No | ||
| 67 | IL11 | 17981 | 6556 | 0.005 | 0.4161 | No | ||
| 68 | RHOA | 8624 4409 4410 | 6567 | 0.005 | 0.4157 | No | ||
| 69 | SYT6 | 13437 7042 12040 | 6621 | 0.005 | 0.4131 | No | ||
| 70 | SOX10 | 22211 | 6685 | 0.004 | 0.4099 | No | ||
| 71 | BMF | 5074 | 6826 | 0.004 | 0.4024 | No | ||
| 72 | CLSTN2 | 19033 | 7257 | 0.003 | 0.3793 | No | ||
| 73 | CALD1 | 4273 8463 | 7286 | 0.003 | 0.3779 | No | ||
| 74 | CAV3 | 17336 | 7287 | 0.003 | 0.3780 | No | ||
| 75 | LTBP3 | 5008 23785 9288 | 7352 | 0.003 | 0.3746 | No | ||
| 76 | LRCH4 | 10544 6090 | 7362 | 0.003 | 0.3743 | No | ||
| 77 | DAB2IP | 12808 2692 | 7511 | 0.002 | 0.3664 | No | ||
| 78 | USP1 | 10497 10498 6051 | 7548 | 0.002 | 0.3645 | No | ||
| 79 | TGFB1I1 | 5742 10159 | 7673 | 0.002 | 0.3579 | No | ||
| 80 | SUMO2 | 1293 9319 | 7838 | 0.002 | 0.3491 | No | ||
| 81 | COL1A1 | 8771 | 7861 | 0.002 | 0.3479 | No | ||
| 82 | TUFT1 | 1847 15253 | 8001 | 0.001 | 0.3405 | No | ||
| 83 | ESM1 | 21560 | 8099 | 0.001 | 0.3352 | No | ||
| 84 | CSF2 | 20454 | 8345 | 0.001 | 0.3220 | No | ||
| 85 | RKHD3 | 18199 | 8478 | 0.000 | 0.3149 | No | ||
| 86 | LY6H | 6244 22256 10732 | 8586 | 0.000 | 0.3091 | No | ||
| 87 | HIF3A | 11989 3784 2303 | 8606 | 0.000 | 0.3080 | No | ||
| 88 | KCTD15 | 17864 | 9012 | -0.001 | 0.2861 | No | ||
| 89 | RTN1 | 4177 2099 | 9134 | -0.001 | 0.2796 | No | ||
| 90 | COX8C | 21175 | 9198 | -0.001 | 0.2762 | No | ||
| 91 | HDAC9 | 21084 2131 | 9290 | -0.001 | 0.2713 | No | ||
| 92 | FBXO24 | 12918 3649 | 9423 | -0.002 | 0.2643 | No | ||
| 93 | SLITRK5 | 21939 | 9587 | -0.002 | 0.2555 | No | ||
| 94 | FOXN1 | 20337 | 10052 | -0.003 | 0.2305 | No | ||
| 95 | ZIC2 | 21927 | 10130 | -0.003 | 0.2264 | No | ||
| 96 | PAX6 | 5223 | 10163 | -0.003 | 0.2249 | No | ||
| 97 | D4ST1 | 14907 | 10206 | -0.003 | 0.2227 | No | ||
| 98 | TCF7L2 | 10048 5646 | 10213 | -0.003 | 0.2226 | No | ||
| 99 | NOPE | 19407 | 10414 | -0.004 | 0.2119 | No | ||
| 100 | LIN28 | 15723 | 10416 | -0.004 | 0.2120 | No | ||
| 101 | B4GALT2 | 11990 | 10453 | -0.004 | 0.2103 | No | ||
| 102 | DDR1 | 1625 23000 | 10510 | -0.004 | 0.2074 | No | ||
| 103 | CALM3 | 8682 | 10582 | -0.004 | 0.2038 | No | ||
| 104 | COL1A2 | 8772 | 10779 | -0.005 | 0.1933 | No | ||
| 105 | PRELP | 13833 | 10987 | -0.005 | 0.1824 | No | ||
| 106 | NNAT | 2764 5180 | 10991 | -0.005 | 0.1824 | No | ||
| 107 | GUCA2B | 15773 | 11071 | -0.006 | 0.1784 | No | ||
| 108 | CNN1 | 8759 | 11209 | -0.006 | 0.1713 | No | ||
| 109 | SLC41A1 | 8269 | 11260 | -0.006 | 0.1688 | No | ||
| 110 | LIM2 | 18282 | 11353 | -0.006 | 0.1641 | No | ||
| 111 | NGFB | 15476 | 11384 | -0.006 | 0.1628 | No | ||
| 112 | PSMA5 | 6464 | 11691 | -0.007 | 0.1465 | No | ||
| 113 | RGMA | 18213 | 11888 | -0.008 | 0.1363 | No | ||
| 114 | DHH | 4629 | 11950 | -0.008 | 0.1333 | No | ||
| 115 | HEBP2 | 19812 | 11977 | -0.008 | 0.1323 | No | ||
| 116 | CDK5R2 | 14220 | 11986 | -0.009 | 0.1323 | No | ||
| 117 | VCP | 11254 | 12000 | -0.009 | 0.1319 | No | ||
| 118 | EGFL6 | 24006 | 12051 | -0.009 | 0.1296 | No | ||
| 119 | KIF1C | 4951 | 12162 | -0.009 | 0.1241 | No | ||
| 120 | IGFBP6 | 9161 | 12243 | -0.010 | 0.1202 | No | ||
| 121 | HOXC5 | 22340 | 12284 | -0.010 | 0.1185 | No | ||
| 122 | ODC1 | 9500 | 12373 | -0.010 | 0.1141 | No | ||
| 123 | GREM1 | 14485 | 12374 | -0.010 | 0.1146 | No | ||
| 124 | ELAVL2 | 15839 | 12384 | -0.010 | 0.1146 | No | ||
| 125 | DHRS3 | 15997 | 12392 | -0.010 | 0.1147 | No | ||
| 126 | DSG3 | 8866 | 12496 | -0.011 | 0.1096 | No | ||
| 127 | GPC3 | 24151 | 12569 | -0.011 | 0.1062 | No | ||
| 128 | IRS4 | 9183 4926 | 12643 | -0.012 | 0.1027 | No | ||
| 129 | ATP6V1E2 | 22880 | 12759 | -0.012 | 0.0971 | No | ||
| 130 | MAP1A | 5124 | 12820 | -0.013 | 0.0944 | No | ||
| 131 | GATA3 | 9004 | 12840 | -0.013 | 0.0939 | No | ||
| 132 | BAP1 | 22057 | 13161 | -0.015 | 0.0772 | No | ||
| 133 | ITGB1BP2 | 24275 | 13356 | -0.016 | 0.0674 | No | ||
| 134 | STAG2 | 5521 | 13520 | -0.018 | 0.0594 | No | ||
| 135 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 13549 | -0.018 | 0.0587 | No | ||
| 136 | WNT3 | 20635 | 13575 | -0.018 | 0.0582 | No | ||
| 137 | VKORC1L1 | 102 7625 | 13600 | -0.018 | 0.0577 | No | ||
| 138 | SLC8A3 | 2065 4293 | 13601 | -0.018 | 0.0585 | No | ||
| 139 | CRTAC1 | 23673 | 13883 | -0.021 | 0.0442 | No | ||
| 140 | ACBD5 | 15105 2751 | 13913 | -0.022 | 0.0436 | No | ||
| 141 | DAB1 | 4594 8834 | 14112 | -0.024 | 0.0340 | No | ||
| 142 | ETV5 | 22630 | 14115 | -0.024 | 0.0350 | No | ||
| 143 | JPH2 | 14364 7210 | 14127 | -0.024 | 0.0355 | No | ||
| 144 | SMOX | 10455 2940 2737 2952 | 14261 | -0.026 | 0.0294 | No | ||
| 145 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 14351 | -0.027 | 0.0258 | No | ||
| 146 | KIF5A | 9222 | 14389 | -0.028 | 0.0251 | No | ||
| 147 | COL11A2 | 23286 | 14435 | -0.029 | 0.0240 | No | ||
| 148 | KCNN2 | 8933 1949 | 14570 | -0.031 | 0.0181 | No | ||
| 149 | ANXA8 | 22046 | 14765 | -0.035 | 0.0092 | No | ||
| 150 | NCDN | 15756 2487 6471 | 14856 | -0.038 | 0.0060 | No | ||
| 151 | HOXC8 | 4865 | 14911 | -0.039 | 0.0049 | No | ||
| 152 | NOL4 | 2028 11432 | 14919 | -0.039 | 0.0063 | No | ||
| 153 | TENC1 | 22349 9927 | 14944 | -0.040 | 0.0068 | No | ||
| 154 | DDIT4 | 19762 | 14950 | -0.040 | 0.0083 | No | ||
| 155 | SOCS1 | 4522 | 14964 | -0.040 | 0.0094 | No | ||
| 156 | ICAM4 | 19544 | 15053 | -0.042 | 0.0065 | No | ||
| 157 | PRDM13 | 15933 | 15162 | -0.045 | 0.0027 | No | ||
| 158 | SOX12 | 5478 | 15278 | -0.049 | -0.0013 | No | ||
| 159 | SORT1 | 15452 5475 9847 | 15411 | -0.053 | -0.0061 | No | ||
| 160 | MYBPC3 | 9434 | 15420 | -0.053 | -0.0041 | No | ||
| 161 | ABHD4 | 22021 | 15511 | -0.057 | -0.0064 | No | ||
| 162 | LAMB2 | 9265 | 15724 | -0.066 | -0.0149 | No | ||
| 163 | CABP1 | 16412 | 15725 | -0.066 | -0.0119 | No | ||
| 164 | SFRS7 | 22889 | 16140 | -0.088 | -0.0304 | No | ||
| 165 | FKBP2 | 8972 | 16221 | -0.093 | -0.0305 | No | ||
| 166 | PTOV1 | 13522 | 16345 | -0.101 | -0.0326 | No | ||
| 167 | ARF6 | 21252 | 16656 | -0.127 | -0.0436 | No | ||
| 168 | PAK2 | 10310 5875 | 16761 | -0.137 | -0.0431 | No | ||
| 169 | RBKS | 16568 | 17144 | -0.180 | -0.0557 | No | ||
| 170 | PANK2 | 14838 | 17269 | -0.194 | -0.0537 | No | ||
| 171 | PHF7 | 21890 | 17559 | -0.232 | -0.0588 | No | ||
| 172 | NDUFS4 | 9452 5157 3173 | 17574 | -0.235 | -0.0490 | No | ||
| 173 | MAPKAP1 | 15030 5993 | 17841 | -0.284 | -0.0505 | No | ||
| 174 | CHCHD4 | 17067 | 17857 | -0.289 | -0.0383 | No | ||
| 175 | PPP6C | 7491 | 18000 | -0.323 | -0.0314 | No | ||
| 176 | CHD4 | 8418 17281 4225 | 18201 | -0.422 | -0.0231 | No | ||
| 177 | ENO1 | 8903 | 18309 | -0.494 | -0.0066 | No | ||
| 178 | RAB10 | 2090 21119 | 18334 | -0.514 | 0.0153 | No |