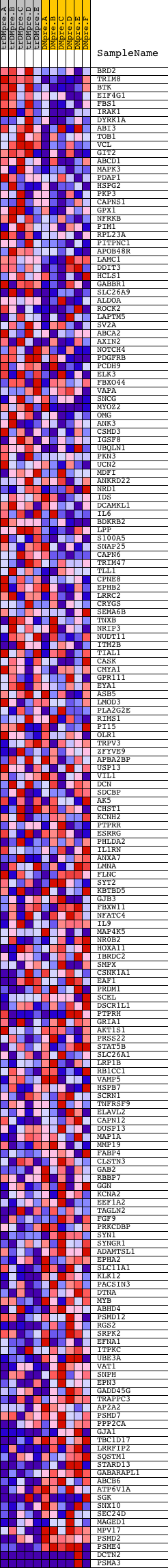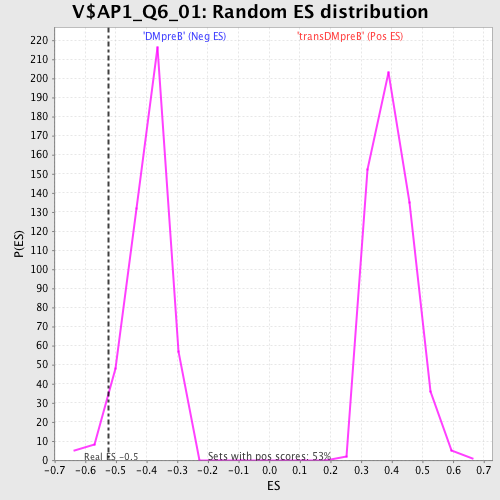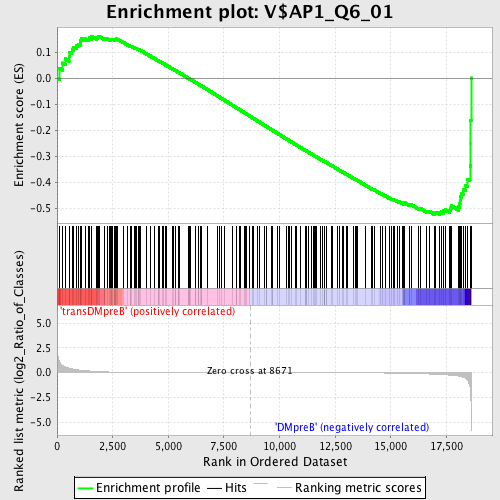
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$AP1_Q6_01 |
| Enrichment Score (ES) | -0.5237724 |
| Normalized Enrichment Score (NES) | -1.3283455 |
| Nominal p-value | 0.030042918 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BRD2 | 4735 8984 7628 | 110 | 1.098 | 0.0377 | No | ||
| 2 | TRIM8 | 23824 | 252 | 0.705 | 0.0580 | No | ||
| 3 | BTK | 24061 | 369 | 0.566 | 0.0742 | No | ||
| 4 | EIF4G1 | 22818 | 537 | 0.453 | 0.0832 | No | ||
| 5 | FBS1 | 18067 | 557 | 0.439 | 0.0996 | No | ||
| 6 | IRAK1 | 4916 | 670 | 0.378 | 0.1085 | No | ||
| 7 | DYRK1A | 4649 | 738 | 0.345 | 0.1186 | No | ||
| 8 | ABI3 | 7314 | 865 | 0.303 | 0.1238 | No | ||
| 9 | TOB1 | 20703 | 942 | 0.278 | 0.1307 | No | ||
| 10 | VCL | 22083 | 1039 | 0.252 | 0.1355 | No | ||
| 11 | GIT2 | 11174 6462 | 1064 | 0.244 | 0.1439 | No | ||
| 12 | ABCD1 | 24305 | 1074 | 0.241 | 0.1530 | No | ||
| 13 | MAPK3 | 6458 11170 | 1268 | 0.198 | 0.1504 | No | ||
| 14 | PDAP1 | 16294 | 1413 | 0.173 | 0.1495 | No | ||
| 15 | HSPG2 | 4884 16023 | 1444 | 0.168 | 0.1545 | No | ||
| 16 | PKP3 | 18016 | 1539 | 0.153 | 0.1555 | No | ||
| 17 | CAPNS1 | 17894 | 1546 | 0.151 | 0.1611 | No | ||
| 18 | GPX1 | 19310 | 1767 | 0.122 | 0.1540 | No | ||
| 19 | NFRKB | 10642 | 1810 | 0.116 | 0.1564 | No | ||
| 20 | PIM1 | 23308 | 1846 | 0.111 | 0.1589 | No | ||
| 21 | RPL23A | 11193 | 1885 | 0.106 | 0.1610 | No | ||
| 22 | PITPNC1 | 7759 | 2112 | 0.082 | 0.1520 | No | ||
| 23 | APOB48R | 18081 | 2139 | 0.080 | 0.1538 | No | ||
| 24 | LAMC1 | 13805 | 2258 | 0.071 | 0.1502 | No | ||
| 25 | DDIT3 | 19857 | 2355 | 0.065 | 0.1476 | No | ||
| 26 | HCLS1 | 22770 | 2414 | 0.061 | 0.1469 | No | ||
| 27 | GABBR1 | 1519 7034 | 2424 | 0.061 | 0.1488 | No | ||
| 28 | SLC26A9 | 11560 980 4067 | 2459 | 0.058 | 0.1493 | No | ||
| 29 | ALDOA | 8572 | 2505 | 0.056 | 0.1490 | No | ||
| 30 | ROCK2 | 21309 5387 | 2573 | 0.052 | 0.1475 | No | ||
| 31 | LAPTM5 | 16061 | 2582 | 0.052 | 0.1491 | No | ||
| 32 | SV2A | 15498 12250 | 2637 | 0.049 | 0.1481 | No | ||
| 33 | ABCA2 | 15083 | 2641 | 0.049 | 0.1499 | No | ||
| 34 | AXIN2 | 20618 | 2667 | 0.048 | 0.1504 | No | ||
| 35 | NOTCH4 | 23277 9475 | 2734 | 0.045 | 0.1486 | No | ||
| 36 | PDGFRB | 23539 | 2973 | 0.037 | 0.1372 | No | ||
| 37 | PCDH9 | 21736 | 3168 | 0.032 | 0.1280 | No | ||
| 38 | ELK3 | 4663 94 3388 | 3305 | 0.029 | 0.1217 | No | ||
| 39 | FBXO44 | 15675 | 3311 | 0.029 | 0.1226 | No | ||
| 40 | VAPA | 22908 | 3345 | 0.028 | 0.1219 | No | ||
| 41 | SNCG | 21878 | 3464 | 0.026 | 0.1166 | No | ||
| 42 | MYOZ2 | 15174 | 3537 | 0.024 | 0.1136 | No | ||
| 43 | OMG | 5209 | 3572 | 0.024 | 0.1127 | No | ||
| 44 | ANK3 | 8591 3445 3304 3381 | 3640 | 0.023 | 0.1100 | No | ||
| 45 | CSMD3 | 10735 7938 | 3666 | 0.022 | 0.1095 | No | ||
| 46 | IGSF8 | 14048 | 3686 | 0.022 | 0.1094 | No | ||
| 47 | UBQLN1 | 3237 3209 3248 3291 | 3751 | 0.021 | 0.1067 | No | ||
| 48 | PKN3 | 15057 | 4028 | 0.018 | 0.0925 | No | ||
| 49 | UCN2 | 10308 | 4177 | 0.016 | 0.0851 | No | ||
| 50 | MDFI | 22950 | 4360 | 0.014 | 0.0758 | No | ||
| 51 | ANKRD22 | 6970 | 4545 | 0.013 | 0.0663 | No | ||
| 52 | NRD1 | 16140 | 4546 | 0.013 | 0.0668 | No | ||
| 53 | IDS | 9142 2617 | 4614 | 0.013 | 0.0637 | No | ||
| 54 | DCAMKL1 | 8838 4600 | 4716 | 0.012 | 0.0587 | No | ||
| 55 | IL6 | 16895 | 4767 | 0.012 | 0.0564 | No | ||
| 56 | BDKRB2 | 21163 | 4889 | 0.011 | 0.0503 | No | ||
| 57 | LPP | 5570 | 4898 | 0.011 | 0.0503 | No | ||
| 58 | S100A5 | 15529 | 5180 | 0.009 | 0.0355 | No | ||
| 59 | SNAP25 | 5466 | 5247 | 0.009 | 0.0322 | No | ||
| 60 | CAPN6 | 24041 | 5300 | 0.009 | 0.0298 | No | ||
| 61 | TRIM47 | 10130 | 5333 | 0.009 | 0.0284 | No | ||
| 62 | TLL1 | 10188 | 5457 | 0.008 | 0.0220 | No | ||
| 63 | CPNE8 | 22156 | 5464 | 0.008 | 0.0220 | No | ||
| 64 | EPHB2 | 4675 2440 8910 | 5507 | 0.008 | 0.0201 | No | ||
| 65 | LRRC2 | 13189 | 5892 | 0.007 | -0.0005 | No | ||
| 66 | CRYGS | 22628 | 5899 | 0.007 | -0.0006 | No | ||
| 67 | SEMA6B | 1587 22932 | 5952 | 0.006 | -0.0031 | No | ||
| 68 | TNXB | 1557 8174 878 8175 | 5971 | 0.006 | -0.0038 | No | ||
| 69 | NRIP3 | 17679 | 6012 | 0.006 | -0.0058 | No | ||
| 70 | NUDT11 | 24387 | 6220 | 0.006 | -0.0168 | No | ||
| 71 | ITM2B | 9192 4935 21753 | 6223 | 0.006 | -0.0167 | No | ||
| 72 | TIAL1 | 996 5753 | 6368 | 0.005 | -0.0243 | No | ||
| 73 | CASK | 24184 2618 2604 | 6374 | 0.005 | -0.0243 | No | ||
| 74 | CMYA1 | 18966 | 6436 | 0.005 | -0.0274 | No | ||
| 75 | GPR111 | 564 | 6475 | 0.005 | -0.0293 | No | ||
| 76 | EYA1 | 4695 4061 | 6502 | 0.005 | -0.0305 | No | ||
| 77 | ASB5 | 13322 | 6751 | 0.004 | -0.0438 | No | ||
| 78 | LMOD3 | 17059 | 7209 | 0.003 | -0.0685 | No | ||
| 79 | PLA2G2E | 16016 | 7319 | 0.003 | -0.0743 | No | ||
| 80 | RIMS1 | 8575 | 7371 | 0.003 | -0.0769 | No | ||
| 81 | PI15 | 13586 | 7508 | 0.002 | -0.0842 | No | ||
| 82 | OLR1 | 8427 4233 | 7897 | 0.002 | -0.1052 | No | ||
| 83 | TRPV3 | 20788 | 8057 | 0.001 | -0.1138 | No | ||
| 84 | ZFYVE9 | 10501 2449 | 8058 | 0.001 | -0.1137 | No | ||
| 85 | APBA2BP | 14385 | 8199 | 0.001 | -0.1213 | No | ||
| 86 | USP13 | 13047 | 8247 | 0.001 | -0.1238 | No | ||
| 87 | VIL1 | 14224 | 8434 | 0.000 | -0.1339 | No | ||
| 88 | DCN | 19897 4601 8840 3366 | 8451 | 0.000 | -0.1347 | No | ||
| 89 | SDCBP | 2339 6998 6999 | 8509 | 0.000 | -0.1378 | No | ||
| 90 | AK5 | 6042 | 8636 | 0.000 | -0.1446 | No | ||
| 91 | CHST1 | 13375 | 8773 | -0.000 | -0.1520 | No | ||
| 92 | KCNH2 | 16592 | 8828 | -0.000 | -0.1549 | No | ||
| 93 | PTPRR | 3436 19876 | 9014 | -0.001 | -0.1649 | No | ||
| 94 | ESRRG | 6447 | 9087 | -0.001 | -0.1688 | No | ||
| 95 | PHLDA2 | 17545 | 9338 | -0.001 | -0.1823 | No | ||
| 96 | IL1RN | 15096 | 9398 | -0.002 | -0.1854 | No | ||
| 97 | ANXA7 | 21908 | 9401 | -0.002 | -0.1855 | No | ||
| 98 | LMNA | 4999 15290 1778 9280 | 9618 | -0.002 | -0.1971 | No | ||
| 99 | FLNC | 4302 8510 | 9669 | -0.002 | -0.1998 | No | ||
| 100 | SYT2 | 14122 | 9696 | -0.002 | -0.2011 | No | ||
| 101 | KBTBD5 | 13020 | 9926 | -0.003 | -0.2134 | No | ||
| 102 | GJB3 | 15754 | 10016 | -0.003 | -0.2181 | No | ||
| 103 | FBXW11 | 20926 | 10329 | -0.004 | -0.2349 | No | ||
| 104 | NFATC4 | 22002 | 10385 | -0.004 | -0.2377 | No | ||
| 105 | IL9 | 21444 | 10435 | -0.004 | -0.2402 | No | ||
| 106 | MAP4K5 | 11853 21048 2101 2142 | 10533 | -0.004 | -0.2453 | No | ||
| 107 | NR0B2 | 16050 | 10729 | -0.005 | -0.2557 | No | ||
| 108 | HOXA11 | 17146 | 10774 | -0.005 | -0.2579 | No | ||
| 109 | IBRDC2 | 5747 | 10920 | -0.005 | -0.2655 | No | ||
| 110 | SMPX | 24220 | 11161 | -0.006 | -0.2783 | No | ||
| 111 | CSNK1A1 | 8204 | 11164 | -0.006 | -0.2782 | No | ||
| 112 | EAF1 | 3634 22055 | 11213 | -0.006 | -0.2806 | No | ||
| 113 | PRDM1 | 19775 3337 | 11218 | -0.006 | -0.2805 | No | ||
| 114 | SCEL | 21942 | 11292 | -0.006 | -0.2843 | No | ||
| 115 | DSCR1L1 | 23231 | 11415 | -0.007 | -0.2906 | No | ||
| 116 | PTPRH | 11861 | 11532 | -0.007 | -0.2966 | No | ||
| 117 | GRIA1 | 20877 4801 9040 | 11556 | -0.007 | -0.2976 | No | ||
| 118 | AKT1S1 | 12557 | 11631 | -0.007 | -0.3013 | No | ||
| 119 | PRSS22 | 12905 | 11639 | -0.007 | -0.3014 | No | ||
| 120 | STAT5B | 20222 | 11851 | -0.008 | -0.3126 | No | ||
| 121 | SLC26A1 | 16446 | 11915 | -0.008 | -0.3156 | No | ||
| 122 | LRP1B | 8249 | 11996 | -0.009 | -0.3196 | No | ||
| 123 | RB1CC1 | 4486 14299 | 12016 | -0.009 | -0.3203 | No | ||
| 124 | VAMP5 | 17118 | 12038 | -0.009 | -0.3211 | No | ||
| 125 | HSPB7 | 16005 | 12112 | -0.009 | -0.3247 | No | ||
| 126 | SCRN1 | 7649 12840 | 12113 | -0.009 | -0.3244 | No | ||
| 127 | TNFRSF9 | 10208 5785 | 12340 | -0.010 | -0.3362 | No | ||
| 128 | ELAVL2 | 15839 | 12384 | -0.010 | -0.3382 | No | ||
| 129 | CAPN12 | 18314 | 12620 | -0.012 | -0.3504 | No | ||
| 130 | DUSP13 | 21904 | 12678 | -0.012 | -0.3531 | No | ||
| 131 | MAP1A | 5124 | 12820 | -0.013 | -0.3602 | No | ||
| 132 | MMP19 | 7191 | 12852 | -0.013 | -0.3614 | No | ||
| 133 | FABP4 | 8601 | 13011 | -0.014 | -0.3694 | No | ||
| 134 | CLSTN3 | 17006 | 13047 | -0.014 | -0.3707 | No | ||
| 135 | GAB2 | 1821 18184 2025 | 13305 | -0.016 | -0.3841 | No | ||
| 136 | RBBP7 | 10887 6366 10886 | 13425 | -0.017 | -0.3898 | No | ||
| 137 | GGN | 2472 1171 18312 | 13456 | -0.017 | -0.3908 | No | ||
| 138 | KCNA2 | 15458 | 13523 | -0.018 | -0.3937 | No | ||
| 139 | EEF1A2 | 8880 14309 | 13854 | -0.021 | -0.4107 | No | ||
| 140 | TAGLN2 | 14043 | 14148 | -0.024 | -0.4257 | No | ||
| 141 | FGF9 | 8966 | 14196 | -0.025 | -0.4272 | No | ||
| 142 | PRKCDBP | 17706 | 14252 | -0.026 | -0.4292 | No | ||
| 143 | SYN1 | 24175 5558 | 14519 | -0.030 | -0.4424 | No | ||
| 144 | SYNGR1 | 22420 2290 | 14633 | -0.033 | -0.4472 | No | ||
| 145 | ADAMTSL1 | 16181 | 14749 | -0.035 | -0.4521 | No | ||
| 146 | EPHA2 | 16006 | 14931 | -0.040 | -0.4603 | No | ||
| 147 | SLC11A1 | 5189 9483 5190 | 15049 | -0.042 | -0.4650 | No | ||
| 148 | KLK12 | 18274 | 15105 | -0.044 | -0.4662 | No | ||
| 149 | PACSIN3 | 14948 8149 2776 2665 | 15173 | -0.046 | -0.4681 | No | ||
| 150 | DTNA | 8870 4647 23611 1991 | 15315 | -0.050 | -0.4737 | No | ||
| 151 | MYB | 3344 3355 19806 | 15381 | -0.052 | -0.4752 | No | ||
| 152 | ABHD4 | 22021 | 15511 | -0.057 | -0.4800 | No | ||
| 153 | PSMD12 | 20621 | 15587 | -0.061 | -0.4816 | No | ||
| 154 | RGS2 | 5379 | 15602 | -0.061 | -0.4800 | No | ||
| 155 | SRPK2 | 5513 | 15615 | -0.062 | -0.4782 | No | ||
| 156 | EFNA1 | 15279 | 15822 | -0.071 | -0.4865 | No | ||
| 157 | ITPKC | 17919 | 15852 | -0.072 | -0.4852 | No | ||
| 158 | UBE3A | 1209 1084 1830 | 15942 | -0.077 | -0.4870 | No | ||
| 159 | VAT1 | 11253 | 16241 | -0.095 | -0.4994 | No | ||
| 160 | SNPH | 14395 | 16353 | -0.102 | -0.5013 | No | ||
| 161 | EPN3 | 20291 | 16620 | -0.124 | -0.5108 | No | ||
| 162 | GADD45G | 21637 | 16736 | -0.135 | -0.5117 | No | ||
| 163 | TRAPPC3 | 6553 16080 2469 11305 | 16959 | -0.160 | -0.5174 | Yes | ||
| 164 | AP2A2 | 18009 | 17026 | -0.168 | -0.5143 | Yes | ||
| 165 | PSMD7 | 18752 3825 | 17181 | -0.183 | -0.5154 | Yes | ||
| 166 | PPP2CA | 20890 | 17280 | -0.195 | -0.5130 | Yes | ||
| 167 | GJA1 | 20023 | 17380 | -0.207 | -0.5101 | Yes | ||
| 168 | TBC1D17 | 6121 | 17450 | -0.218 | -0.5052 | Yes | ||
| 169 | LRRFIP2 | 3024 3071 19285 | 17658 | -0.247 | -0.5066 | Yes | ||
| 170 | SQSTM1 | 9517 | 17701 | -0.253 | -0.4988 | Yes | ||
| 171 | STARD13 | 10833 | 17709 | -0.255 | -0.4891 | Yes | ||
| 172 | GABARAPL1 | 17268 | 18045 | -0.337 | -0.4938 | Yes | ||
| 173 | ABCB6 | 13162 | 18097 | -0.356 | -0.4825 | Yes | ||
| 174 | ATP6V1A | 8638 | 18111 | -0.359 | -0.4689 | Yes | ||
| 175 | SGK | 9809 3419 | 18118 | -0.363 | -0.4548 | Yes | ||
| 176 | SNX10 | 17445 | 18161 | -0.394 | -0.4414 | Yes | ||
| 177 | SEC24D | 15429 | 18262 | -0.461 | -0.4285 | Yes | ||
| 178 | MAGED1 | 24108 | 18348 | -0.533 | -0.4119 | Yes | ||
| 179 | MPV17 | 9407 3505 3543 | 18447 | -0.731 | -0.3882 | Yes | ||
| 180 | PSMD2 | 10137 5724 | 18559 | -1.443 | -0.3369 | Yes | ||
| 181 | PSME4 | 8339 20934 | 18587 | -2.192 | -0.2513 | Yes | ||
| 182 | DCTN2 | 7635 12812 | 18588 | -2.219 | -0.1631 | Yes | ||
| 183 | PSMA3 | 9632 5298 | 18610 | -4.143 | 0.0003 | Yes |