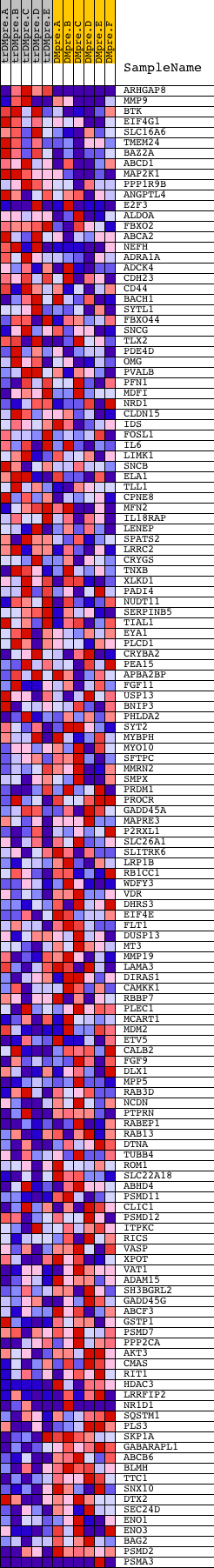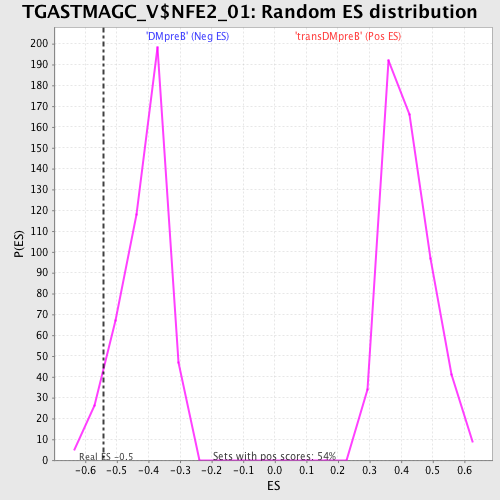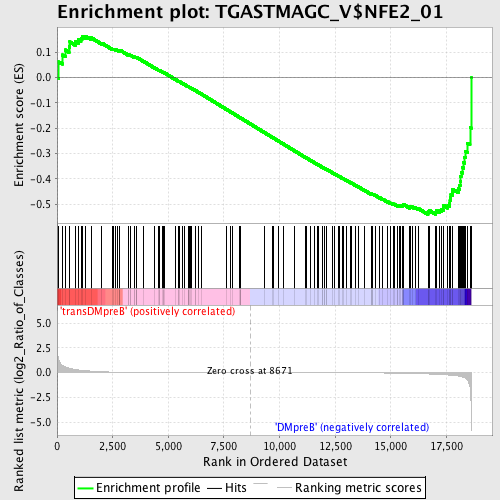
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | TGASTMAGC_V$NFE2_01 |
| Enrichment Score (ES) | -0.54141355 |
| Normalized Enrichment Score (NES) | -1.3135614 |
| Nominal p-value | 0.04338395 |
| FDR q-value | 0.96761715 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARHGAP8 | 4268 2196 | 73 | 1.369 | 0.0621 | No | ||
| 2 | MMP9 | 14732 | 230 | 0.746 | 0.0897 | No | ||
| 3 | BTK | 24061 | 369 | 0.566 | 0.1095 | No | ||
| 4 | EIF4G1 | 22818 | 537 | 0.453 | 0.1223 | No | ||
| 5 | SLC16A6 | 4186 | 573 | 0.431 | 0.1412 | No | ||
| 6 | TMEM24 | 19150 | 814 | 0.318 | 0.1436 | No | ||
| 7 | BAZ2A | 4371 | 950 | 0.276 | 0.1496 | No | ||
| 8 | ABCD1 | 24305 | 1074 | 0.241 | 0.1546 | No | ||
| 9 | MAP2K1 | 19082 | 1120 | 0.227 | 0.1631 | No | ||
| 10 | PPP1R9B | 20698 | 1275 | 0.195 | 0.1642 | No | ||
| 11 | ANGPTL4 | 1504 7178 | 1529 | 0.155 | 0.1580 | No | ||
| 12 | E2F3 | 21500 8874 | 2002 | 0.092 | 0.1368 | No | ||
| 13 | ALDOA | 8572 | 2505 | 0.056 | 0.1124 | No | ||
| 14 | FBXO2 | 15993 | 2552 | 0.053 | 0.1124 | No | ||
| 15 | ABCA2 | 15083 | 2641 | 0.049 | 0.1100 | No | ||
| 16 | NEFH | 11724 | 2719 | 0.046 | 0.1081 | No | ||
| 17 | ADRA1A | 2850 8560 | 2791 | 0.043 | 0.1063 | No | ||
| 18 | ADCK4 | 13364 | 2817 | 0.043 | 0.1070 | No | ||
| 19 | CDH23 | 3369 19759 3442 | 2820 | 0.042 | 0.1090 | No | ||
| 20 | CD44 | 2881 2847 2893 4499 2724 | 3206 | 0.031 | 0.0896 | No | ||
| 21 | BACH1 | 8647 | 3207 | 0.031 | 0.0911 | No | ||
| 22 | SYTL1 | 15728 | 3310 | 0.029 | 0.0870 | No | ||
| 23 | FBXO44 | 15675 | 3311 | 0.029 | 0.0884 | No | ||
| 24 | SNCG | 21878 | 3464 | 0.026 | 0.0814 | No | ||
| 25 | TLX2 | 5773 1165 | 3478 | 0.025 | 0.0819 | No | ||
| 26 | PDE4D | 10722 6235 | 3548 | 0.024 | 0.0793 | No | ||
| 27 | OMG | 5209 | 3572 | 0.024 | 0.0792 | No | ||
| 28 | PVALB | 22225 | 3579 | 0.024 | 0.0801 | No | ||
| 29 | PFN1 | 9555 | 3886 | 0.019 | 0.0644 | No | ||
| 30 | MDFI | 22950 | 4360 | 0.014 | 0.0395 | No | ||
| 31 | NRD1 | 16140 | 4546 | 0.013 | 0.0301 | No | ||
| 32 | CLDN15 | 16667 3572 | 4579 | 0.013 | 0.0290 | No | ||
| 33 | IDS | 9142 2617 | 4614 | 0.013 | 0.0278 | No | ||
| 34 | FOSL1 | 23779 | 4732 | 0.012 | 0.0220 | No | ||
| 35 | IL6 | 16895 | 4767 | 0.012 | 0.0208 | No | ||
| 36 | LIMK1 | 16350 | 4780 | 0.012 | 0.0207 | No | ||
| 37 | SNCB | 21456 | 4816 | 0.011 | 0.0193 | No | ||
| 38 | ELA1 | 22127 | 5323 | 0.009 | -0.0077 | No | ||
| 39 | TLL1 | 10188 | 5457 | 0.008 | -0.0145 | No | ||
| 40 | CPNE8 | 22156 | 5464 | 0.008 | -0.0144 | No | ||
| 41 | MFN2 | 15678 2417 | 5491 | 0.008 | -0.0154 | No | ||
| 42 | IL18RAP | 14262 | 5631 | 0.008 | -0.0226 | No | ||
| 43 | LENEP | 12181 | 5729 | 0.007 | -0.0275 | No | ||
| 44 | SPATS2 | 13044 | 5884 | 0.007 | -0.0355 | No | ||
| 45 | LRRC2 | 13189 | 5892 | 0.007 | -0.0355 | No | ||
| 46 | CRYGS | 22628 | 5899 | 0.007 | -0.0355 | No | ||
| 47 | TNXB | 1557 8174 878 8175 | 5971 | 0.006 | -0.0391 | No | ||
| 48 | XLKD1 | 17675 | 5996 | 0.006 | -0.0401 | No | ||
| 49 | PADI4 | 2424 9547 | 6036 | 0.006 | -0.0419 | No | ||
| 50 | NUDT11 | 24387 | 6220 | 0.006 | -0.0515 | No | ||
| 51 | SERPINB5 | 9861 | 6238 | 0.006 | -0.0522 | No | ||
| 52 | TIAL1 | 996 5753 | 6368 | 0.005 | -0.0589 | No | ||
| 53 | EYA1 | 4695 4061 | 6502 | 0.005 | -0.0659 | No | ||
| 54 | PLCD1 | 18972 | 7594 | 0.002 | -0.1248 | No | ||
| 55 | CRYBA2 | 13920 | 7792 | 0.002 | -0.1354 | No | ||
| 56 | PEA15 | 13753 | 7887 | 0.002 | -0.1404 | No | ||
| 57 | APBA2BP | 14385 | 8199 | 0.001 | -0.1572 | No | ||
| 58 | FGF11 | 20383 | 8220 | 0.001 | -0.1582 | No | ||
| 59 | USP13 | 13047 | 8247 | 0.001 | -0.1596 | No | ||
| 60 | BNIP3 | 17581 | 8252 | 0.001 | -0.1598 | No | ||
| 61 | PHLDA2 | 17545 | 9338 | -0.001 | -0.2184 | No | ||
| 62 | SYT2 | 14122 | 9696 | -0.002 | -0.2376 | No | ||
| 63 | MYBPH | 14130 | 9737 | -0.002 | -0.2397 | No | ||
| 64 | MYO10 | 2184 5141 | 9967 | -0.003 | -0.2519 | No | ||
| 65 | SFTPC | 21759 | 10165 | -0.003 | -0.2624 | No | ||
| 66 | MMRN2 | 4194 | 10683 | -0.005 | -0.2902 | No | ||
| 67 | SMPX | 24220 | 11161 | -0.006 | -0.3157 | No | ||
| 68 | PRDM1 | 19775 3337 | 11218 | -0.006 | -0.3185 | No | ||
| 69 | PROCR | 14772 | 11410 | -0.007 | -0.3285 | No | ||
| 70 | GADD45A | 17129 | 11554 | -0.007 | -0.3359 | No | ||
| 71 | MAPRE3 | 8301 | 11720 | -0.008 | -0.3445 | No | ||
| 72 | P2RXL1 | 5218 9521 | 11747 | -0.008 | -0.3455 | No | ||
| 73 | SLC26A1 | 16446 | 11915 | -0.008 | -0.3542 | No | ||
| 74 | SLITRK6 | 21724 | 11948 | -0.008 | -0.3555 | No | ||
| 75 | LRP1B | 8249 | 11996 | -0.009 | -0.3576 | No | ||
| 76 | RB1CC1 | 4486 14299 | 12016 | -0.009 | -0.3582 | No | ||
| 77 | WDFY3 | 7789 3607 16462 | 12104 | -0.009 | -0.3625 | No | ||
| 78 | VDR | 5852 10285 | 12119 | -0.009 | -0.3628 | No | ||
| 79 | DHRS3 | 15997 | 12392 | -0.010 | -0.3770 | No | ||
| 80 | EIF4E | 15403 1827 8890 | 12487 | -0.011 | -0.3816 | No | ||
| 81 | FLT1 | 3483 16287 | 12654 | -0.012 | -0.3900 | No | ||
| 82 | DUSP13 | 21904 | 12678 | -0.012 | -0.3907 | No | ||
| 83 | MT3 | 9414 | 12816 | -0.013 | -0.3975 | No | ||
| 84 | MMP19 | 7191 | 12852 | -0.013 | -0.3988 | No | ||
| 85 | LAMA3 | 23620 2009 | 13003 | -0.014 | -0.4062 | No | ||
| 86 | DIRAS1 | 9915 | 13202 | -0.015 | -0.4162 | No | ||
| 87 | CAMKK1 | 20793 | 13231 | -0.015 | -0.4170 | No | ||
| 88 | RBBP7 | 10887 6366 10886 | 13425 | -0.017 | -0.4267 | No | ||
| 89 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 13549 | -0.018 | -0.4324 | No | ||
| 90 | MCART1 | 10492 | 13837 | -0.021 | -0.4470 | No | ||
| 91 | MDM2 | 19620 3327 | 14114 | -0.024 | -0.4608 | No | ||
| 92 | ETV5 | 22630 | 14115 | -0.024 | -0.4596 | No | ||
| 93 | CALB2 | 8680 18748 | 14158 | -0.024 | -0.4607 | No | ||
| 94 | FGF9 | 8966 | 14196 | -0.025 | -0.4615 | No | ||
| 95 | DLX1 | 14988 | 14308 | -0.027 | -0.4662 | No | ||
| 96 | MPP5 | 21230 7078 | 14511 | -0.030 | -0.4757 | No | ||
| 97 | RAB3D | 19205 | 14624 | -0.033 | -0.4802 | No | ||
| 98 | NCDN | 15756 2487 6471 | 14856 | -0.038 | -0.4909 | No | ||
| 99 | PTPRN | 13914 | 14987 | -0.041 | -0.4959 | No | ||
| 100 | RABEP1 | 12016 1344 | 15108 | -0.044 | -0.5003 | No | ||
| 101 | RAB13 | 15535 | 15142 | -0.045 | -0.4999 | No | ||
| 102 | DTNA | 8870 4647 23611 1991 | 15315 | -0.050 | -0.5068 | No | ||
| 103 | TUBB4 | 22917 | 15320 | -0.050 | -0.5047 | No | ||
| 104 | ROM1 | 23751 | 15367 | -0.051 | -0.5047 | No | ||
| 105 | SLC22A18 | 18000 | 15453 | -0.054 | -0.5066 | No | ||
| 106 | ABHD4 | 22021 | 15511 | -0.057 | -0.5070 | No | ||
| 107 | PSMD11 | 12772 7600 | 15523 | -0.058 | -0.5048 | No | ||
| 108 | CLIC1 | 23262 | 15549 | -0.059 | -0.5033 | No | ||
| 109 | PSMD12 | 20621 | 15587 | -0.061 | -0.5024 | No | ||
| 110 | ITPKC | 17919 | 15852 | -0.072 | -0.5132 | No | ||
| 111 | RICS | 19523 | 15861 | -0.073 | -0.5101 | No | ||
| 112 | VASP | 5847 | 15954 | -0.078 | -0.5114 | No | ||
| 113 | XPOT | 13090 | 16087 | -0.084 | -0.5145 | No | ||
| 114 | VAT1 | 11253 | 16241 | -0.095 | -0.5181 | No | ||
| 115 | ADAM15 | 15275 | 16672 | -0.129 | -0.5352 | Yes | ||
| 116 | SH3BGRL2 | 5601 | 16680 | -0.130 | -0.5293 | Yes | ||
| 117 | GADD45G | 21637 | 16736 | -0.135 | -0.5258 | Yes | ||
| 118 | ABCF3 | 6578 | 17015 | -0.166 | -0.5328 | Yes | ||
| 119 | GSTP1 | 9051 4812 4811 | 17055 | -0.171 | -0.5267 | Yes | ||
| 120 | PSMD7 | 18752 3825 | 17181 | -0.183 | -0.5246 | Yes | ||
| 121 | PPP2CA | 20890 | 17280 | -0.195 | -0.5205 | Yes | ||
| 122 | AKT3 | 13739 982 | 17357 | -0.204 | -0.5147 | Yes | ||
| 123 | CMAS | 17245 | 17362 | -0.205 | -0.5051 | Yes | ||
| 124 | RIT1 | 5382 | 17565 | -0.234 | -0.5047 | Yes | ||
| 125 | HDAC3 | 4844 | 17648 | -0.246 | -0.4973 | Yes | ||
| 126 | LRRFIP2 | 3024 3071 19285 | 17658 | -0.247 | -0.4859 | Yes | ||
| 127 | NR1D1 | 4650 20258 949 | 17684 | -0.251 | -0.4751 | Yes | ||
| 128 | SQSTM1 | 9517 | 17701 | -0.253 | -0.4638 | Yes | ||
| 129 | PLS3 | 8332 | 17767 | -0.268 | -0.4544 | Yes | ||
| 130 | SKP1A | 20889 | 17782 | -0.271 | -0.4420 | Yes | ||
| 131 | GABARAPL1 | 17268 | 18045 | -0.337 | -0.4400 | Yes | ||
| 132 | ABCB6 | 13162 | 18097 | -0.356 | -0.4256 | Yes | ||
| 133 | BLMH | 20769 | 18117 | -0.363 | -0.4091 | Yes | ||
| 134 | TTC1 | 7342 | 18139 | -0.380 | -0.3919 | Yes | ||
| 135 | SNX10 | 17445 | 18161 | -0.394 | -0.3740 | Yes | ||
| 136 | DTX2 | 16674 3488 | 18229 | -0.441 | -0.3563 | Yes | ||
| 137 | SEC24D | 15429 | 18262 | -0.461 | -0.3358 | Yes | ||
| 138 | ENO1 | 8903 | 18309 | -0.494 | -0.3145 | Yes | ||
| 139 | ENO3 | 8905 | 18359 | -0.547 | -0.2908 | Yes | ||
| 140 | BAG2 | 13985 | 18444 | -0.725 | -0.2603 | Yes | ||
| 141 | PSMD2 | 10137 5724 | 18559 | -1.443 | -0.1969 | Yes | ||
| 142 | PSMA3 | 9632 5298 | 18610 | -4.143 | 0.0003 | Yes |