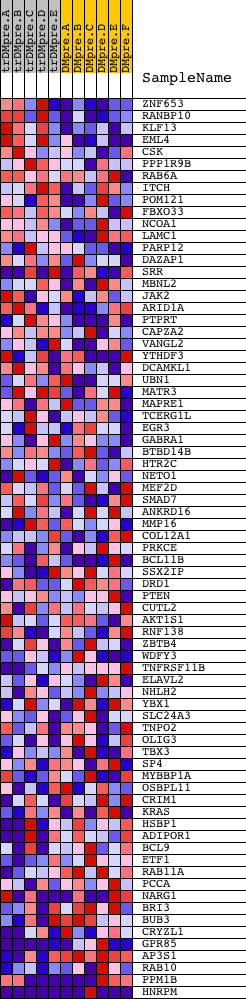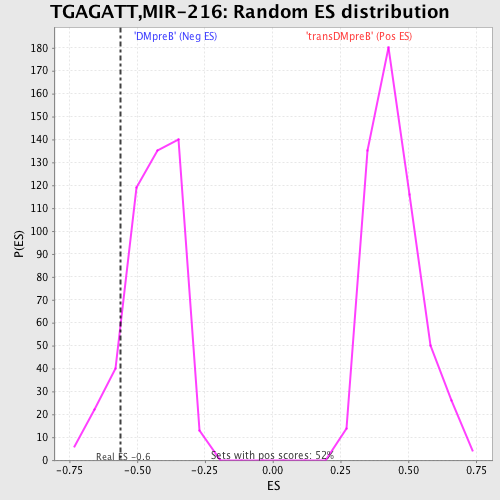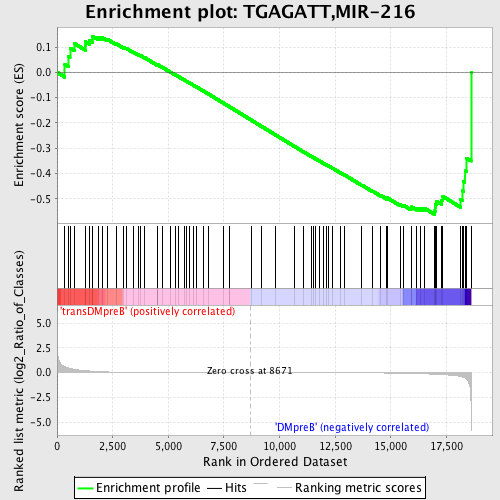
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | TGAGATT,MIR-216 |
| Enrichment Score (ES) | -0.56117994 |
| Normalized Enrichment Score (NES) | -1.2643734 |
| Nominal p-value | 0.11368421 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ZNF653 | 11459 | 350 | 0.583 | 0.0312 | No | ||
| 2 | RANBP10 | 18764 | 503 | 0.472 | 0.0635 | No | ||
| 3 | KLF13 | 17807 | 580 | 0.428 | 0.0961 | No | ||
| 4 | EML4 | 8122 | 773 | 0.332 | 0.1142 | No | ||
| 5 | CSK | 8805 | 1270 | 0.197 | 0.1043 | No | ||
| 6 | PPP1R9B | 20698 | 1275 | 0.195 | 0.1208 | No | ||
| 7 | RAB6A | 18173 3905 | 1469 | 0.163 | 0.1244 | No | ||
| 8 | ITCH | 4928 9185 | 1589 | 0.147 | 0.1306 | No | ||
| 9 | POM121 | 16343 | 1592 | 0.146 | 0.1430 | No | ||
| 10 | FBXO33 | 528 | 1857 | 0.110 | 0.1382 | No | ||
| 11 | NCOA1 | 5154 | 2028 | 0.089 | 0.1367 | No | ||
| 12 | LAMC1 | 13805 | 2258 | 0.071 | 0.1304 | No | ||
| 13 | PARP12 | 6335 | 2663 | 0.048 | 0.1127 | No | ||
| 14 | DAZAP1 | 19945 | 2992 | 0.037 | 0.0981 | No | ||
| 15 | SRR | 6566 11328 | 3103 | 0.034 | 0.0951 | No | ||
| 16 | MBNL2 | 21932 | 3427 | 0.026 | 0.0799 | No | ||
| 17 | JAK2 | 23893 9197 3706 | 3648 | 0.023 | 0.0700 | No | ||
| 18 | ARID1A | 8214 2501 13554 | 3753 | 0.021 | 0.0662 | No | ||
| 19 | PTPRT | 5333 | 3937 | 0.019 | 0.0580 | No | ||
| 20 | CAPZA2 | 8688 991 | 4491 | 0.013 | 0.0293 | No | ||
| 21 | VANGL2 | 4109 8220 | 4496 | 0.013 | 0.0302 | No | ||
| 22 | YTHDF3 | 15630 10469 6019 | 4520 | 0.013 | 0.0301 | No | ||
| 23 | DCAMKL1 | 8838 4600 | 4716 | 0.012 | 0.0206 | No | ||
| 24 | UBN1 | 22867 | 5095 | 0.010 | 0.0011 | No | ||
| 25 | MATR3 | 2043 5081 | 5305 | 0.009 | -0.0094 | No | ||
| 26 | MAPRE1 | 4652 | 5436 | 0.008 | -0.0157 | No | ||
| 27 | TCERG1L | 7690 | 5717 | 0.007 | -0.0302 | No | ||
| 28 | EGR3 | 4656 | 5831 | 0.007 | -0.0357 | No | ||
| 29 | GABRA1 | 20492 | 5938 | 0.006 | -0.0409 | No | ||
| 30 | BTBD14B | 7344 7343 | 6123 | 0.006 | -0.0503 | No | ||
| 31 | HTR2C | 24230 | 6285 | 0.005 | -0.0585 | No | ||
| 32 | NETO1 | 6372 1970 | 6559 | 0.005 | -0.0728 | No | ||
| 33 | MEF2D | 9379 | 6793 | 0.004 | -0.0850 | No | ||
| 34 | SMAD7 | 23513 | 7464 | 0.002 | -0.1209 | No | ||
| 35 | ANKRD16 | 6804 | 7746 | 0.002 | -0.1359 | No | ||
| 36 | MMP16 | 5109 | 7752 | 0.002 | -0.1361 | No | ||
| 37 | COL12A1 | 3143 19054 | 8716 | -0.000 | -0.1880 | No | ||
| 38 | PRKCE | 9575 | 9168 | -0.001 | -0.2122 | No | ||
| 39 | BCL11B | 7190 | 9826 | -0.003 | -0.2474 | No | ||
| 40 | SSX2IP | 13617 | 10653 | -0.005 | -0.2916 | No | ||
| 41 | DRD1 | 4638 | 11085 | -0.006 | -0.3143 | No | ||
| 42 | PTEN | 5305 | 11437 | -0.007 | -0.3327 | No | ||
| 43 | CUTL2 | 13110 4578 | 11540 | -0.007 | -0.3376 | No | ||
| 44 | AKT1S1 | 12557 | 11631 | -0.007 | -0.3418 | No | ||
| 45 | RNF138 | 2006 23613 2029 2037 | 11802 | -0.008 | -0.3503 | No | ||
| 46 | ZBTB4 | 13267 | 11975 | -0.008 | -0.3589 | No | ||
| 47 | WDFY3 | 7789 3607 16462 | 12104 | -0.009 | -0.3650 | No | ||
| 48 | TNFRSF11B | 22296 | 12187 | -0.009 | -0.3686 | No | ||
| 49 | ELAVL2 | 15839 | 12384 | -0.010 | -0.3783 | No | ||
| 50 | NHLH2 | 9462 | 12748 | -0.012 | -0.3969 | No | ||
| 51 | YBX1 | 5929 2389 | 12935 | -0.013 | -0.4057 | No | ||
| 52 | SLC24A3 | 14818 | 13703 | -0.019 | -0.4454 | No | ||
| 53 | TNPO2 | 5608 | 14188 | -0.025 | -0.4694 | No | ||
| 54 | OLIG3 | 20084 | 14542 | -0.031 | -0.4858 | No | ||
| 55 | TBX3 | 16723 | 14823 | -0.037 | -0.4977 | No | ||
| 56 | SP4 | 20972 | 14839 | -0.037 | -0.4953 | No | ||
| 57 | MYBBP1A | 5217 1232 5216 | 15431 | -0.054 | -0.5226 | No | ||
| 58 | OSBPL11 | 22781 | 15566 | -0.060 | -0.5247 | No | ||
| 59 | CRIM1 | 403 | 15913 | -0.075 | -0.5369 | No | ||
| 60 | KRAS | 9247 | 15933 | -0.076 | -0.5314 | No | ||
| 61 | HSBP1 | 18451 | 16161 | -0.090 | -0.5359 | No | ||
| 62 | ADIPOR1 | 14126 | 16342 | -0.101 | -0.5370 | No | ||
| 63 | BCL9 | 15232 | 16512 | -0.114 | -0.5363 | No | ||
| 64 | ETF1 | 23467 | 16974 | -0.162 | -0.5473 | Yes | ||
| 65 | RAB11A | 7013 | 16990 | -0.164 | -0.5340 | Yes | ||
| 66 | PCCA | 21926 | 17001 | -0.165 | -0.5205 | Yes | ||
| 67 | NARG1 | 7934 917 | 17058 | -0.171 | -0.5088 | Yes | ||
| 68 | BRI3 | 15101 | 17287 | -0.196 | -0.5043 | Yes | ||
| 69 | BUB3 | 18045 | 17339 | -0.202 | -0.4898 | Yes | ||
| 70 | CRYZL1 | 1644 12385 | 18131 | -0.371 | -0.5006 | Yes | ||
| 71 | GPR85 | 7227 1048 | 18235 | -0.444 | -0.4681 | Yes | ||
| 72 | AP3S1 | 8604 | 18273 | -0.467 | -0.4300 | Yes | ||
| 73 | RAB10 | 2090 21119 | 18334 | -0.514 | -0.3892 | Yes | ||
| 74 | PPM1B | 5280 1567 5281 | 18410 | -0.627 | -0.3395 | Yes | ||
| 75 | HNRPM | 1511 13370 | 18607 | -4.087 | 0.0005 | Yes |