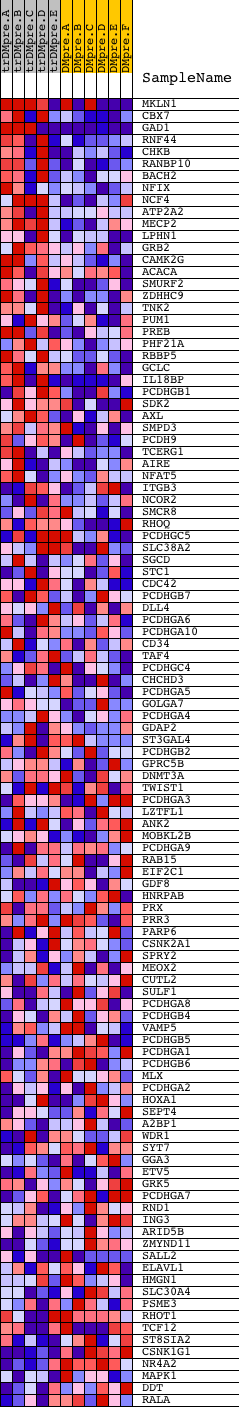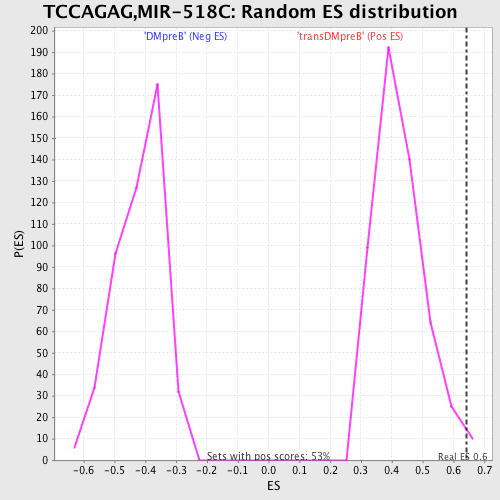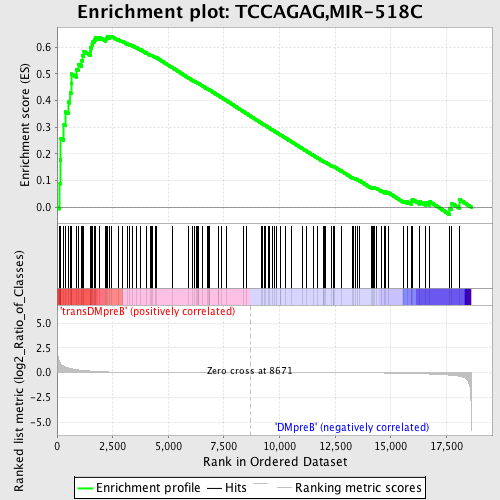
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | TCCAGAG,MIR-518C |
| Enrichment Score (ES) | 0.64195275 |
| Normalized Enrichment Score (NES) | 1.5097893 |
| Nominal p-value | 0.013207547 |
| FDR q-value | 0.2983152 |
| FWER p-Value | 0.956 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MKLN1 | 6584 11339 | 127 | 1.016 | 0.0872 | Yes | ||
| 2 | CBX7 | 11973 | 134 | 0.988 | 0.1784 | Yes | ||
| 3 | GAD1 | 14993 2690 | 168 | 0.889 | 0.2590 | Yes | ||
| 4 | RNF44 | 8364 | 306 | 0.631 | 0.3101 | Yes | ||
| 5 | CHKB | 4518 | 383 | 0.556 | 0.3575 | Yes | ||
| 6 | RANBP10 | 18764 | 503 | 0.472 | 0.3948 | Yes | ||
| 7 | BACH2 | 8648 | 579 | 0.429 | 0.4305 | Yes | ||
| 8 | NFIX | 9458 | 630 | 0.396 | 0.4645 | Yes | ||
| 9 | NCF4 | 22435 | 641 | 0.389 | 0.5001 | Yes | ||
| 10 | ATP2A2 | 4421 3481 | 863 | 0.303 | 0.5162 | Yes | ||
| 11 | MECP2 | 5088 | 951 | 0.275 | 0.5370 | Yes | ||
| 12 | LPHN1 | 6850 174 | 1097 | 0.232 | 0.5507 | Yes | ||
| 13 | GRB2 | 20149 | 1140 | 0.224 | 0.5692 | Yes | ||
| 14 | CAMK2G | 21905 | 1204 | 0.210 | 0.5853 | Yes | ||
| 15 | ACACA | 309 4209 | 1489 | 0.160 | 0.5848 | Yes | ||
| 16 | SMURF2 | 185 | 1499 | 0.159 | 0.5990 | Yes | ||
| 17 | ZDHHC9 | 24163 | 1567 | 0.149 | 0.6093 | Yes | ||
| 18 | TNK2 | 22784 | 1609 | 0.143 | 0.6203 | Yes | ||
| 19 | PUM1 | 8160 | 1663 | 0.135 | 0.6300 | Yes | ||
| 20 | PREB | 6948 3628 | 1728 | 0.125 | 0.6381 | Yes | ||
| 21 | PHF21A | 14943 | 1913 | 0.103 | 0.6377 | Yes | ||
| 22 | RBBP5 | 5620 5619 | 2181 | 0.077 | 0.6304 | Yes | ||
| 23 | GCLC | 19374 | 2223 | 0.074 | 0.6350 | Yes | ||
| 24 | IL18BP | 17730 | 2247 | 0.072 | 0.6404 | Yes | ||
| 25 | PCDHGB1 | 13532 8207 | 2368 | 0.064 | 0.6398 | Yes | ||
| 26 | SDK2 | 6208 20162 | 2432 | 0.060 | 0.6420 | Yes | ||
| 27 | AXL | 17922 | 2751 | 0.045 | 0.6289 | No | ||
| 28 | SMPD3 | 18757 | 2918 | 0.039 | 0.6236 | No | ||
| 29 | PCDH9 | 21736 | 3168 | 0.032 | 0.6132 | No | ||
| 30 | TCERG1 | 12075 2027 7068 | 3265 | 0.030 | 0.6107 | No | ||
| 31 | AIRE | 19716 3386 | 3404 | 0.027 | 0.6057 | No | ||
| 32 | NFAT5 | 3921 7037 12036 | 3565 | 0.024 | 0.5993 | No | ||
| 33 | ITGB3 | 20631 | 3749 | 0.021 | 0.5914 | No | ||
| 34 | NCOR2 | 9835 16367 | 4002 | 0.018 | 0.5794 | No | ||
| 35 | SMCR8 | 53 | 4191 | 0.016 | 0.5708 | No | ||
| 36 | RHOQ | 23142 | 4237 | 0.015 | 0.5698 | No | ||
| 37 | PCDHGC5 | 13540 8208 | 4270 | 0.015 | 0.5694 | No | ||
| 38 | SLC38A2 | 22149 2219 | 4432 | 0.014 | 0.5620 | No | ||
| 39 | SGCD | 1425 10779 | 4478 | 0.014 | 0.5609 | No | ||
| 40 | STC1 | 21974 | 5176 | 0.009 | 0.5241 | No | ||
| 41 | CDC42 | 4503 8722 4504 2465 | 5905 | 0.007 | 0.4854 | No | ||
| 42 | PCDHGB7 | 13537 | 6076 | 0.006 | 0.4767 | No | ||
| 43 | DLL4 | 7040 | 6155 | 0.006 | 0.4731 | No | ||
| 44 | PCDHGA6 | 13546 | 6254 | 0.006 | 0.4683 | No | ||
| 45 | PCDHGA10 | 13550 | 6294 | 0.005 | 0.4667 | No | ||
| 46 | CD34 | 14006 | 6354 | 0.005 | 0.4640 | No | ||
| 47 | TAF4 | 14319 | 6365 | 0.005 | 0.4639 | No | ||
| 48 | PCDHGC4 | 13539 | 6537 | 0.005 | 0.4551 | No | ||
| 49 | CHCHD3 | 17188 | 6777 | 0.004 | 0.4426 | No | ||
| 50 | PCDHGA5 | 13545 | 6808 | 0.004 | 0.4413 | No | ||
| 51 | GOLGA7 | 12188 | 6830 | 0.004 | 0.4406 | No | ||
| 52 | PCDHGA4 | 13544 | 6866 | 0.004 | 0.4390 | No | ||
| 53 | GDAP2 | 15481 | 7246 | 0.003 | 0.4188 | No | ||
| 54 | ST3GAL4 | 9816 | 7267 | 0.003 | 0.4180 | No | ||
| 55 | PCDHGB2 | 13533 | 7402 | 0.003 | 0.4110 | No | ||
| 56 | GPRC5B | 17662 | 7591 | 0.002 | 0.4011 | No | ||
| 57 | DNMT3A | 2167 21330 | 8386 | 0.001 | 0.3582 | No | ||
| 58 | TWIST1 | 21291 | 8501 | 0.000 | 0.3521 | No | ||
| 59 | PCDHGA3 | 13543 | 9200 | -0.001 | 0.3145 | No | ||
| 60 | LZTFL1 | 8210 | 9235 | -0.001 | 0.3128 | No | ||
| 61 | ANK2 | 4275 | 9300 | -0.001 | 0.3094 | No | ||
| 62 | MOBKL2B | 15922 | 9344 | -0.001 | 0.3072 | No | ||
| 63 | PCDHGA9 | 13549 | 9513 | -0.002 | 0.2983 | No | ||
| 64 | RAB15 | 21035 | 9563 | -0.002 | 0.2959 | No | ||
| 65 | EIF2C1 | 10672 | 9701 | -0.002 | 0.2887 | No | ||
| 66 | GDF8 | 9411 | 9791 | -0.002 | 0.2841 | No | ||
| 67 | HNRPAB | 9104 | 9870 | -0.003 | 0.2801 | No | ||
| 68 | PRX | 3729 9628 | 10033 | -0.003 | 0.2716 | No | ||
| 69 | PRR3 | 22995 | 10060 | -0.003 | 0.2705 | No | ||
| 70 | PARP6 | 19419 2979 3156 | 10277 | -0.004 | 0.2592 | No | ||
| 71 | CSNK2A1 | 14797 | 10540 | -0.004 | 0.2454 | No | ||
| 72 | SPRY2 | 21725 | 11036 | -0.006 | 0.2192 | No | ||
| 73 | MEOX2 | 21285 | 11191 | -0.006 | 0.2114 | No | ||
| 74 | CUTL2 | 13110 4578 | 11540 | -0.007 | 0.1933 | No | ||
| 75 | SULF1 | 6285 | 11714 | -0.008 | 0.1846 | No | ||
| 76 | PCDHGA8 | 13548 | 11960 | -0.008 | 0.1722 | No | ||
| 77 | PCDHGB4 | 13534 | 11987 | -0.009 | 0.1715 | No | ||
| 78 | VAMP5 | 17118 | 12038 | -0.009 | 0.1696 | No | ||
| 79 | PCDHGB5 | 13535 | 12079 | -0.009 | 0.1683 | No | ||
| 80 | PCDHGA1 | 13541 | 12348 | -0.010 | 0.1548 | No | ||
| 81 | PCDHGB6 | 13536 | 12401 | -0.010 | 0.1529 | No | ||
| 82 | MLX | 958 1417 | 12403 | -0.010 | 0.1538 | No | ||
| 83 | PCDHGA2 | 13542 | 12447 | -0.011 | 0.1525 | No | ||
| 84 | HOXA1 | 1005 17152 | 12766 | -0.012 | 0.1364 | No | ||
| 85 | SEPT4 | 20713 | 13278 | -0.016 | 0.1103 | No | ||
| 86 | A2BP1 | 11212 1652 1689 | 13329 | -0.016 | 0.1091 | No | ||
| 87 | WDR1 | 16544 3592 | 13396 | -0.017 | 0.1070 | No | ||
| 88 | SYT7 | 3670 3698 23932 | 13488 | -0.017 | 0.1037 | No | ||
| 89 | GGA3 | 1206 6435 | 13607 | -0.018 | 0.0991 | No | ||
| 90 | ETV5 | 22630 | 14115 | -0.024 | 0.0739 | No | ||
| 91 | GRK5 | 9036 23814 | 14155 | -0.024 | 0.0740 | No | ||
| 92 | PCDHGA7 | 13547 | 14214 | -0.025 | 0.0732 | No | ||
| 93 | RND1 | 5868 | 14256 | -0.026 | 0.0734 | No | ||
| 94 | ING3 | 17514 1019 | 14342 | -0.027 | 0.0713 | No | ||
| 95 | ARID5B | 7728 | 14587 | -0.032 | 0.0611 | No | ||
| 96 | ZMYND11 | 12364 3269 3286 3223 | 14737 | -0.035 | 0.0563 | No | ||
| 97 | SALL2 | 21840 | 14743 | -0.035 | 0.0592 | No | ||
| 98 | ELAVL1 | 4888 | 14882 | -0.038 | 0.0553 | No | ||
| 99 | HMGN1 | 4856 | 15582 | -0.060 | 0.0231 | No | ||
| 100 | SLC30A4 | 14450 | 15743 | -0.067 | 0.0207 | No | ||
| 101 | PSME3 | 20657 | 15912 | -0.075 | 0.0186 | No | ||
| 102 | RHOT1 | 12227 | 15920 | -0.076 | 0.0252 | No | ||
| 103 | TCF12 | 10044 3125 | 15972 | -0.078 | 0.0297 | No | ||
| 104 | ST8SIA2 | 17797 | 16309 | -0.099 | 0.0208 | No | ||
| 105 | CSNK1G1 | 3057 5671 19397 | 16576 | -0.120 | 0.0175 | No | ||
| 106 | NR4A2 | 5203 | 16755 | -0.136 | 0.0205 | No | ||
| 107 | MAPK1 | 1642 11167 | 17642 | -0.244 | -0.0048 | No | ||
| 108 | DDT | 19732 | 17744 | -0.262 | 0.0141 | No | ||
| 109 | RALA | 21541 | 18100 | -0.356 | 0.0279 | No |