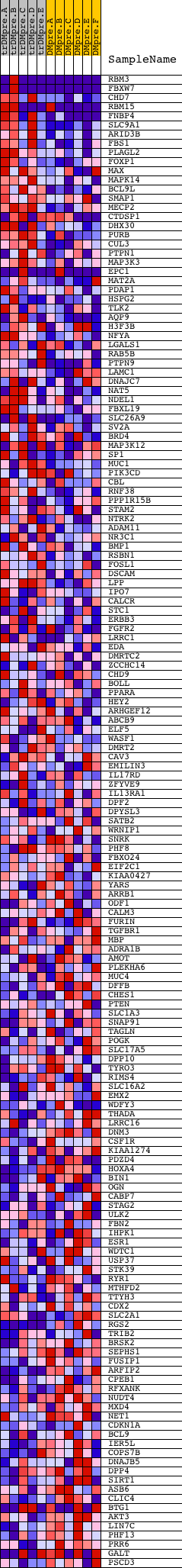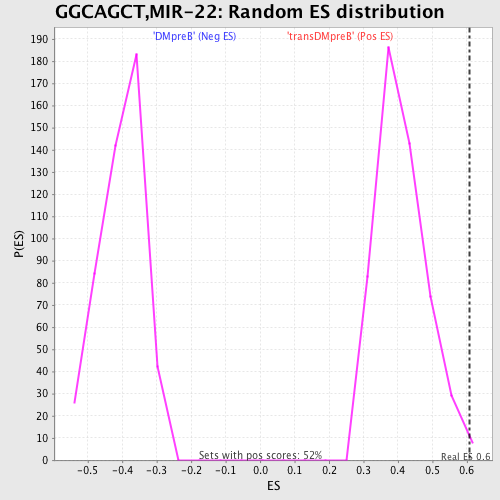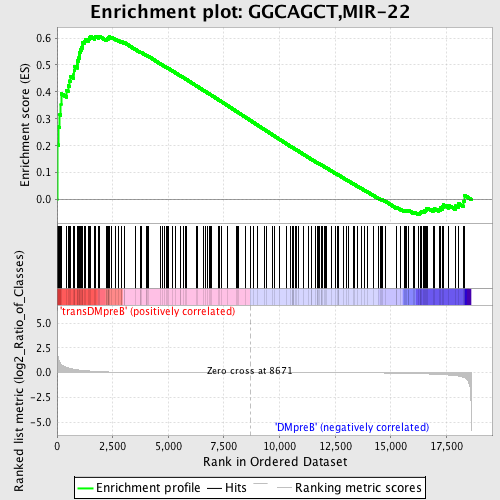
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | GGCAGCT,MIR-22 |
| Enrichment Score (ES) | 0.6070739 |
| Normalized Enrichment Score (NES) | 1.4834682 |
| Nominal p-value | 0.009560229 |
| FDR q-value | 0.30664667 |
| FWER p-Value | 0.988 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RBM3 | 5366 2625 | 2 | 4.390 | 0.2049 | Yes | ||
| 2 | FBXW7 | 1805 11928 | 65 | 1.463 | 0.2699 | Yes | ||
| 3 | CHD7 | 16278 | 122 | 1.038 | 0.3153 | Yes | ||
| 4 | RBM15 | 5215 15207 | 173 | 0.875 | 0.3535 | Yes | ||
| 5 | FNBP4 | 14956 2671 | 177 | 0.869 | 0.3939 | Yes | ||
| 6 | SLC9A1 | 16053 | 425 | 0.520 | 0.4048 | Yes | ||
| 7 | ARID3B | 12111 | 498 | 0.475 | 0.4231 | Yes | ||
| 8 | FBS1 | 18067 | 557 | 0.439 | 0.4405 | Yes | ||
| 9 | PLAGL2 | 7055 | 592 | 0.421 | 0.4583 | Yes | ||
| 10 | FOXP1 | 4242 | 755 | 0.338 | 0.4653 | Yes | ||
| 11 | MAX | 21034 | 759 | 0.337 | 0.4809 | Yes | ||
| 12 | MAPK14 | 23313 | 774 | 0.331 | 0.4956 | Yes | ||
| 13 | BCL9L | 19475 | 907 | 0.290 | 0.5020 | Yes | ||
| 14 | SMAP1 | 13603 8268 | 913 | 0.289 | 0.5152 | Yes | ||
| 15 | MECP2 | 5088 | 951 | 0.275 | 0.5261 | Yes | ||
| 16 | CTDSP1 | 4121 10407 | 998 | 0.263 | 0.5359 | Yes | ||
| 17 | DHX30 | 18989 | 1011 | 0.258 | 0.5473 | Yes | ||
| 18 | PURB | 5335 | 1055 | 0.247 | 0.5565 | Yes | ||
| 19 | CUL3 | 6470 | 1102 | 0.231 | 0.5648 | Yes | ||
| 20 | PTPN1 | 5325 | 1144 | 0.223 | 0.5729 | Yes | ||
| 21 | MAP3K3 | 20626 | 1150 | 0.222 | 0.5830 | Yes | ||
| 22 | EPC1 | 2039 8907 2033 | 1241 | 0.204 | 0.5877 | Yes | ||
| 23 | MAT2A | 10553 10554 6099 | 1282 | 0.194 | 0.5945 | Yes | ||
| 24 | PDAP1 | 16294 | 1413 | 0.173 | 0.5956 | Yes | ||
| 25 | HSPG2 | 4884 16023 | 1444 | 0.168 | 0.6018 | Yes | ||
| 26 | TLK2 | 20628 | 1494 | 0.159 | 0.6066 | Yes | ||
| 27 | AQP9 | 7218 | 1698 | 0.129 | 0.6016 | Yes | ||
| 28 | H3F3B | 4839 | 1708 | 0.128 | 0.6071 | Yes | ||
| 29 | NFYA | 5172 | 1855 | 0.110 | 0.6043 | No | ||
| 30 | LGALS1 | 22429 | 1914 | 0.103 | 0.6060 | No | ||
| 31 | RAB5B | 359 19591 3430 | 2221 | 0.074 | 0.5928 | No | ||
| 32 | PTPN9 | 19434 | 2235 | 0.073 | 0.5955 | No | ||
| 33 | LAMC1 | 13805 | 2258 | 0.071 | 0.5976 | No | ||
| 34 | DNAJC7 | 20227 | 2293 | 0.068 | 0.5990 | No | ||
| 35 | NAT5 | 18601 7496 12597 | 2311 | 0.067 | 0.6012 | No | ||
| 36 | NDEL1 | 20399 | 2330 | 0.066 | 0.6033 | No | ||
| 37 | FBXL19 | 6134 | 2360 | 0.064 | 0.6047 | No | ||
| 38 | SLC26A9 | 11560 980 4067 | 2459 | 0.058 | 0.6021 | No | ||
| 39 | SV2A | 15498 12250 | 2637 | 0.049 | 0.5948 | No | ||
| 40 | BRD4 | 7164 1609 1628 1598 1552 1601 12176 1630 1501 | 2775 | 0.044 | 0.5895 | No | ||
| 41 | MAP3K12 | 6454 11164 | 2903 | 0.040 | 0.5844 | No | ||
| 42 | SP1 | 9852 | 2910 | 0.040 | 0.5860 | No | ||
| 43 | MUC1 | 1849 1799 1787 24354 | 3007 | 0.036 | 0.5825 | No | ||
| 44 | PIK3CD | 9563 | 3036 | 0.036 | 0.5826 | No | ||
| 45 | CBL | 19154 | 3514 | 0.025 | 0.5579 | No | ||
| 46 | RNF38 | 7862 2328 13115 | 3736 | 0.021 | 0.5469 | No | ||
| 47 | PPP1R15B | 4254 14135 | 3757 | 0.021 | 0.5468 | No | ||
| 48 | STAM2 | 7095 2845 | 3783 | 0.021 | 0.5465 | No | ||
| 49 | NTRK2 | 9491 | 4026 | 0.018 | 0.5342 | No | ||
| 50 | ADAM11 | 4340 1432 | 4038 | 0.018 | 0.5344 | No | ||
| 51 | NR3C1 | 9043 | 4075 | 0.017 | 0.5333 | No | ||
| 52 | BMP1 | 3268 8658 21758 | 4109 | 0.017 | 0.5323 | No | ||
| 53 | RSBN1 | 6035 | 4638 | 0.012 | 0.5042 | No | ||
| 54 | FOSL1 | 23779 | 4732 | 0.012 | 0.4997 | No | ||
| 55 | DSCAM | 1667 4641 22530 | 4842 | 0.011 | 0.4944 | No | ||
| 56 | LPP | 5570 | 4898 | 0.011 | 0.4919 | No | ||
| 57 | IPO7 | 6130 | 4971 | 0.010 | 0.4885 | No | ||
| 58 | CALCR | 17229 | 4995 | 0.010 | 0.4877 | No | ||
| 59 | STC1 | 21974 | 5176 | 0.009 | 0.4784 | No | ||
| 60 | ERBB3 | 19593 | 5341 | 0.009 | 0.4699 | No | ||
| 61 | FGFR2 | 1917 4722 1119 | 5527 | 0.008 | 0.4603 | No | ||
| 62 | LRRC1 | 19059 3129 | 5551 | 0.008 | 0.4594 | No | ||
| 63 | EDA | 24284 | 5685 | 0.007 | 0.4525 | No | ||
| 64 | DMRTC2 | 18344 | 5782 | 0.007 | 0.4477 | No | ||
| 65 | ZCCHC14 | 4731 | 5827 | 0.007 | 0.4456 | No | ||
| 66 | CHD9 | 6784 11553 18531 | 6261 | 0.006 | 0.4224 | No | ||
| 67 | BOLL | 7960 | 6311 | 0.005 | 0.4200 | No | ||
| 68 | PPARA | 22397 | 6579 | 0.005 | 0.4057 | No | ||
| 69 | HEY2 | 19794 | 6654 | 0.004 | 0.4019 | No | ||
| 70 | ARHGEF12 | 19156 | 6770 | 0.004 | 0.3959 | No | ||
| 71 | ABCB9 | 3576 3546 7096 | 6850 | 0.004 | 0.3918 | No | ||
| 72 | ELF5 | 14936 2913 | 6912 | 0.004 | 0.3887 | No | ||
| 73 | WASF1 | 13514 | 6958 | 0.004 | 0.3864 | No | ||
| 74 | DMRT2 | 23897 | 7256 | 0.003 | 0.3704 | No | ||
| 75 | CAV3 | 17336 | 7287 | 0.003 | 0.3689 | No | ||
| 76 | EMILIN3 | 11376 | 7380 | 0.003 | 0.3641 | No | ||
| 77 | IL17RD | 22069 | 7678 | 0.002 | 0.3481 | No | ||
| 78 | ZFYVE9 | 10501 2449 | 8058 | 0.001 | 0.3276 | No | ||
| 79 | IL13RA1 | 24361 | 8122 | 0.001 | 0.3242 | No | ||
| 80 | DPF2 | 9719 | 8147 | 0.001 | 0.3230 | No | ||
| 81 | DPYSL3 | 10252 | 8456 | 0.000 | 0.3063 | No | ||
| 82 | SATB2 | 5604 | 8702 | -0.000 | 0.2930 | No | ||
| 83 | WRNIP1 | 21675 3161 | 8833 | -0.000 | 0.2860 | No | ||
| 84 | SNRK | 5467 5468 9840 | 9022 | -0.001 | 0.2759 | No | ||
| 85 | PHF8 | 6770 | 9306 | -0.001 | 0.2606 | No | ||
| 86 | FBXO24 | 12918 3649 | 9423 | -0.002 | 0.2544 | No | ||
| 87 | EIF2C1 | 10672 | 9701 | -0.002 | 0.2395 | No | ||
| 88 | KIAA0427 | 11228 | 9771 | -0.002 | 0.2358 | No | ||
| 89 | YARS | 16071 | 9979 | -0.003 | 0.2248 | No | ||
| 90 | ARRB1 | 3890 8465 18178 4276 | 10304 | -0.004 | 0.2074 | No | ||
| 91 | ODF1 | 22488 | 10508 | -0.004 | 0.1966 | No | ||
| 92 | CALM3 | 8682 | 10582 | -0.004 | 0.1928 | No | ||
| 93 | FURIN | 9538 | 10584 | -0.004 | 0.1930 | No | ||
| 94 | TGFBR1 | 16213 10165 5745 | 10618 | -0.004 | 0.1914 | No | ||
| 95 | MBP | 23502 | 10641 | -0.005 | 0.1904 | No | ||
| 96 | ADRA1B | 4354 | 10736 | -0.005 | 0.1855 | No | ||
| 97 | AMOT | 11341 24037 6587 | 10754 | -0.005 | 0.1848 | No | ||
| 98 | PLEKHA6 | 10786 6288 | 10830 | -0.005 | 0.1810 | No | ||
| 99 | MUC4 | 8929 1746 | 11054 | -0.006 | 0.1692 | No | ||
| 100 | DFFB | 15654 | 11313 | -0.006 | 0.1555 | No | ||
| 101 | CHES1 | 21007 11200 | 11432 | -0.007 | 0.1494 | No | ||
| 102 | PTEN | 5305 | 11437 | -0.007 | 0.1495 | No | ||
| 103 | SLC1A3 | 22331 | 11606 | -0.007 | 0.1407 | No | ||
| 104 | SNAP91 | 9839 19043 | 11701 | -0.007 | 0.1360 | No | ||
| 105 | TAGLN | 19136 | 11715 | -0.008 | 0.1356 | No | ||
| 106 | POGK | 7739 | 11738 | -0.008 | 0.1348 | No | ||
| 107 | SLC17A5 | 19055 | 11780 | -0.008 | 0.1329 | No | ||
| 108 | DPP10 | 13850 | 11861 | -0.008 | 0.1290 | No | ||
| 109 | TYRO3 | 5811 | 11880 | -0.008 | 0.1284 | No | ||
| 110 | RIMS4 | 14360 | 11909 | -0.008 | 0.1273 | No | ||
| 111 | SLC16A2 | 24086 | 12039 | -0.009 | 0.1207 | No | ||
| 112 | EMX2 | 23806 | 12077 | -0.009 | 0.1191 | No | ||
| 113 | WDFY3 | 7789 3607 16462 | 12104 | -0.009 | 0.1181 | No | ||
| 114 | THADA | 11614 | 12345 | -0.010 | 0.1056 | No | ||
| 115 | LRRC16 | 21515 21514 | 12526 | -0.011 | 0.0963 | No | ||
| 116 | DNM3 | 5948 | 12619 | -0.012 | 0.0919 | No | ||
| 117 | CSF1R | 8800 23540 | 12626 | -0.012 | 0.0921 | No | ||
| 118 | KIAA1274 | 19756 | 12664 | -0.012 | 0.0906 | No | ||
| 119 | PDZD4 | 24133 2550 2610 | 12871 | -0.013 | 0.0801 | No | ||
| 120 | HOXA4 | 17148 | 12989 | -0.014 | 0.0744 | No | ||
| 121 | BIN1 | 6634 2017 | 13112 | -0.014 | 0.0684 | No | ||
| 122 | OGN | 21642 3215 | 13314 | -0.016 | 0.0583 | No | ||
| 123 | CABP7 | 20542 | 13385 | -0.016 | 0.0553 | No | ||
| 124 | STAG2 | 5521 | 13520 | -0.018 | 0.0488 | No | ||
| 125 | ULK2 | 6617 11384 | 13666 | -0.019 | 0.0419 | No | ||
| 126 | FBN2 | 23433 | 13831 | -0.021 | 0.0339 | No | ||
| 127 | IHPK1 | 11336 | 13929 | -0.022 | 0.0297 | No | ||
| 128 | ESR1 | 20097 4685 | 14203 | -0.025 | 0.0161 | No | ||
| 129 | WDTC1 | 10512 | 14445 | -0.029 | 0.0044 | No | ||
| 130 | USP37 | 13923 | 14545 | -0.031 | 0.0005 | No | ||
| 131 | STK39 | 14570 | 14588 | -0.032 | -0.0003 | No | ||
| 132 | RYR1 | 17903 17902 9770 | 14603 | -0.032 | 0.0004 | No | ||
| 133 | MTHFD2 | 17100 | 14773 | -0.036 | -0.0071 | No | ||
| 134 | TTYH3 | 16312 | 15245 | -0.048 | -0.0304 | No | ||
| 135 | CDX2 | 16289 | 15271 | -0.049 | -0.0294 | No | ||
| 136 | SLC2A1 | 16107 | 15445 | -0.054 | -0.0363 | No | ||
| 137 | RGS2 | 5379 | 15602 | -0.061 | -0.0419 | No | ||
| 138 | TRIB2 | 21102 | 15668 | -0.064 | -0.0424 | No | ||
| 139 | BRSK2 | 190 | 15693 | -0.065 | -0.0407 | No | ||
| 140 | SEPHS1 | 15125 | 15789 | -0.069 | -0.0426 | No | ||
| 141 | FUSIP1 | 4715 16036 | 15811 | -0.070 | -0.0405 | No | ||
| 142 | ARFIP2 | 1226 17702 | 16025 | -0.081 | -0.0482 | No | ||
| 143 | CPEB1 | 8778 | 16075 | -0.084 | -0.0470 | No | ||
| 144 | RFXANK | 5378 | 16224 | -0.093 | -0.0506 | No | ||
| 145 | NUDT4 | 19639 | 16311 | -0.099 | -0.0507 | No | ||
| 146 | MXD4 | 9346 | 16339 | -0.101 | -0.0474 | No | ||
| 147 | NET1 | 7100 | 16373 | -0.103 | -0.0444 | No | ||
| 148 | CDKN1A | 4511 8729 | 16471 | -0.111 | -0.0444 | No | ||
| 149 | BCL9 | 15232 | 16512 | -0.114 | -0.0413 | No | ||
| 150 | IER5L | 14627 | 16572 | -0.119 | -0.0389 | No | ||
| 151 | COPS7B | 6502 11222 | 16592 | -0.121 | -0.0343 | No | ||
| 152 | DNAJB5 | 16236 | 16659 | -0.128 | -0.0319 | No | ||
| 153 | DPP4 | 14579 | 16902 | -0.154 | -0.0378 | No | ||
| 154 | SIRT1 | 19745 | 16953 | -0.159 | -0.0331 | No | ||
| 155 | ASB6 | 2728 14625 | 17165 | -0.182 | -0.0360 | No | ||
| 156 | CLIC4 | 11386 11342 6620 11387 | 17225 | -0.188 | -0.0304 | No | ||
| 157 | BTG1 | 4458 4457 8663 | 17328 | -0.200 | -0.0266 | No | ||
| 158 | AKT3 | 13739 982 | 17357 | -0.204 | -0.0186 | No | ||
| 159 | LIN7C | 14925 | 17598 | -0.238 | -0.0205 | No | ||
| 160 | PHF13 | 15659 | 17916 | -0.302 | -0.0235 | No | ||
| 161 | PRR6 | 20409 | 18052 | -0.339 | -0.0150 | No | ||
| 162 | GALT | 16237 2399 | 18280 | -0.472 | -0.0052 | No | ||
| 163 | PSCD3 | 16633 | 18321 | -0.501 | 0.0160 | No |