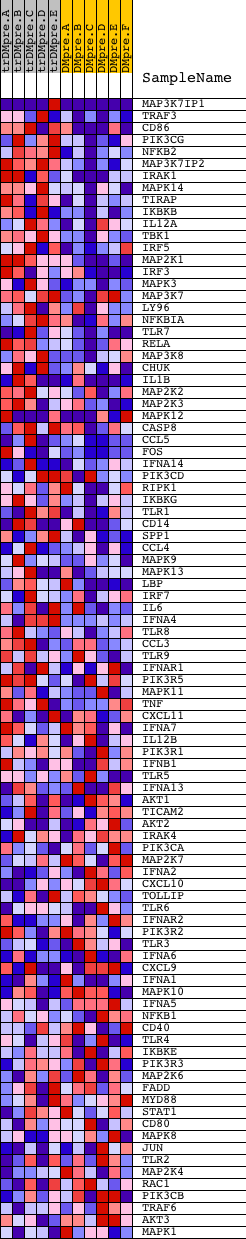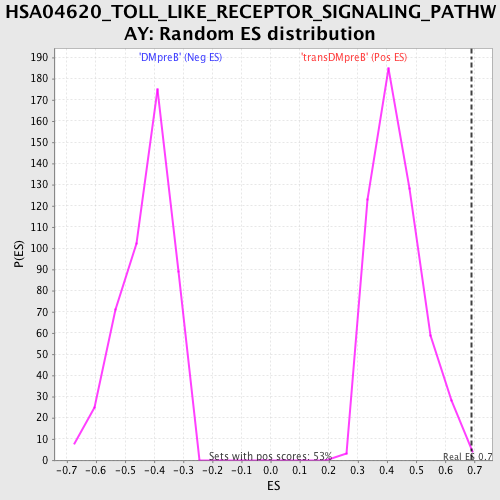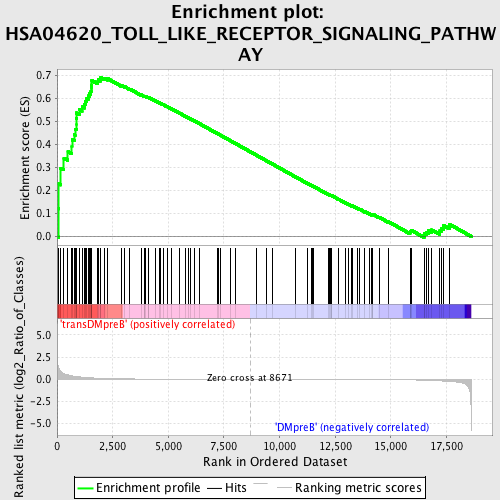
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | HSA04620_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.6891478 |
| Normalized Enrichment Score (NES) | 1.5931277 |
| Nominal p-value | 0.0018867925 |
| FDR q-value | 0.6969804 |
| FWER p-Value | 0.661 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAP3K7IP1 | 2193 2171 22419 | 59 | 1.517 | 0.1200 | Yes | ||
| 2 | TRAF3 | 21147 | 76 | 1.340 | 0.2280 | Yes | ||
| 3 | CD86 | 1660 22601 | 165 | 0.896 | 0.2960 | Yes | ||
| 4 | PIK3CG | 6635 | 307 | 0.628 | 0.3394 | Yes | ||
| 5 | NFKB2 | 23810 | 488 | 0.478 | 0.3685 | Yes | ||
| 6 | MAP3K7IP2 | 19827 | 667 | 0.380 | 0.3898 | Yes | ||
| 7 | IRAK1 | 4916 | 670 | 0.378 | 0.4204 | Yes | ||
| 8 | MAPK14 | 23313 | 774 | 0.331 | 0.4417 | Yes | ||
| 9 | TIRAP | 3075 19187 | 810 | 0.320 | 0.4658 | Yes | ||
| 10 | IKBKB | 4907 | 861 | 0.304 | 0.4878 | Yes | ||
| 11 | IL12A | 4913 | 864 | 0.303 | 0.5123 | Yes | ||
| 12 | TBK1 | 12137 | 878 | 0.299 | 0.5359 | Yes | ||
| 13 | IRF5 | 17502 | 1007 | 0.260 | 0.5500 | Yes | ||
| 14 | MAP2K1 | 19082 | 1120 | 0.227 | 0.5624 | Yes | ||
| 15 | IRF3 | 2149 2188 18255 | 1221 | 0.207 | 0.5738 | Yes | ||
| 16 | MAPK3 | 6458 11170 | 1268 | 0.198 | 0.5874 | Yes | ||
| 17 | MAP3K7 | 16255 | 1335 | 0.182 | 0.5986 | Yes | ||
| 18 | LY96 | 14292 | 1396 | 0.175 | 0.6096 | Yes | ||
| 19 | NFKBIA | 21065 | 1447 | 0.167 | 0.6204 | Yes | ||
| 20 | TLR7 | 24004 | 1517 | 0.156 | 0.6294 | Yes | ||
| 21 | RELA | 23783 | 1532 | 0.154 | 0.6412 | Yes | ||
| 22 | MAP3K8 | 23495 | 1536 | 0.153 | 0.6535 | Yes | ||
| 23 | CHUK | 23665 | 1559 | 0.150 | 0.6645 | Yes | ||
| 24 | IL1B | 14431 | 1560 | 0.150 | 0.6766 | Yes | ||
| 25 | MAP2K2 | 19933 | 1831 | 0.114 | 0.6713 | Yes | ||
| 26 | MAP2K3 | 20856 | 1847 | 0.111 | 0.6795 | Yes | ||
| 27 | MAPK12 | 2182 2215 22164 | 1948 | 0.098 | 0.6821 | Yes | ||
| 28 | CASP8 | 8694 | 1963 | 0.096 | 0.6891 | Yes | ||
| 29 | CCL5 | 20320 | 2144 | 0.080 | 0.6859 | No | ||
| 30 | FOS | 21202 | 2284 | 0.069 | 0.6840 | No | ||
| 31 | IFNA14 | 11917 | 2890 | 0.040 | 0.6546 | No | ||
| 32 | PIK3CD | 9563 | 3036 | 0.036 | 0.6497 | No | ||
| 33 | RIPK1 | 5381 | 3271 | 0.030 | 0.6394 | No | ||
| 34 | IKBKG | 2570 2562 4908 | 3770 | 0.021 | 0.6142 | No | ||
| 35 | TLR1 | 10190 | 3934 | 0.019 | 0.6070 | No | ||
| 36 | CD14 | 23452 | 3948 | 0.019 | 0.6078 | No | ||
| 37 | SPP1 | 5501 | 3974 | 0.018 | 0.6079 | No | ||
| 38 | CCL4 | 1362 20730 | 4096 | 0.017 | 0.6028 | No | ||
| 39 | MAPK9 | 1233 20903 1383 | 4118 | 0.017 | 0.6030 | No | ||
| 40 | MAPK13 | 23314 | 4416 | 0.014 | 0.5881 | No | ||
| 41 | LBP | 14760 | 4598 | 0.013 | 0.5793 | No | ||
| 42 | IRF7 | 12008 | 4629 | 0.013 | 0.5787 | No | ||
| 43 | IL6 | 16895 | 4767 | 0.012 | 0.5723 | No | ||
| 44 | IFNA4 | 9150 | 4940 | 0.011 | 0.5639 | No | ||
| 45 | TLR8 | 9308 | 5147 | 0.010 | 0.5535 | No | ||
| 46 | CCL3 | 20317 | 5519 | 0.008 | 0.5341 | No | ||
| 47 | TLR9 | 19331 3127 | 5787 | 0.007 | 0.5203 | No | ||
| 48 | IFNAR1 | 22703 | 5926 | 0.007 | 0.5134 | No | ||
| 49 | PIK3R5 | 11507 | 6008 | 0.006 | 0.5095 | No | ||
| 50 | MAPK11 | 2264 9618 22163 | 6180 | 0.006 | 0.5008 | No | ||
| 51 | TNF | 23004 | 6407 | 0.005 | 0.4890 | No | ||
| 52 | CXCL11 | 16478 | 7227 | 0.003 | 0.4450 | No | ||
| 53 | IFNA7 | 9152 | 7241 | 0.003 | 0.4445 | No | ||
| 54 | IL12B | 20918 | 7351 | 0.003 | 0.4389 | No | ||
| 55 | PIK3R1 | 3170 | 7781 | 0.002 | 0.4159 | No | ||
| 56 | IFNB1 | 15846 | 8030 | 0.001 | 0.4026 | No | ||
| 57 | TLR5 | 14017 | 8983 | -0.001 | 0.3512 | No | ||
| 58 | IFNA13 | 15844 | 9413 | -0.002 | 0.3282 | No | ||
| 59 | AKT1 | 8568 | 9661 | -0.002 | 0.3150 | No | ||
| 60 | TICAM2 | 23440 | 10724 | -0.005 | 0.2581 | No | ||
| 61 | AKT2 | 4365 4366 | 11243 | -0.006 | 0.2306 | No | ||
| 62 | IRAK4 | 22379 11185 | 11414 | -0.007 | 0.2220 | No | ||
| 63 | PIK3CA | 9562 | 11488 | -0.007 | 0.2186 | No | ||
| 64 | MAP2K7 | 6453 | 11502 | -0.007 | 0.2184 | No | ||
| 65 | IFNA2 | 9149 | 12196 | -0.009 | 0.1818 | No | ||
| 66 | CXCL10 | 16477 | 12249 | -0.010 | 0.1797 | No | ||
| 67 | TOLLIP | 7039 12038 | 12269 | -0.010 | 0.1795 | No | ||
| 68 | TLR6 | 215 16528 | 12328 | -0.010 | 0.1772 | No | ||
| 69 | IFNAR2 | 22705 1699 | 12350 | -0.010 | 0.1769 | No | ||
| 70 | PIK3R2 | 18850 | 12644 | -0.012 | 0.1620 | No | ||
| 71 | TLR3 | 18884 | 12979 | -0.014 | 0.1451 | No | ||
| 72 | IFNA6 | 17749 | 13107 | -0.014 | 0.1394 | No | ||
| 73 | CXCL9 | 5099 | 13252 | -0.015 | 0.1329 | No | ||
| 74 | IFNA1 | 9147 | 13280 | -0.016 | 0.1327 | No | ||
| 75 | MAPK10 | 11169 | 13486 | -0.017 | 0.1230 | No | ||
| 76 | IFNA5 | 9151 | 13605 | -0.018 | 0.1182 | No | ||
| 77 | NFKB1 | 15160 | 13838 | -0.021 | 0.1073 | No | ||
| 78 | CD40 | 10207 2961 2968 5783 | 14028 | -0.023 | 0.0990 | No | ||
| 79 | TLR4 | 2329 10191 5770 | 14147 | -0.024 | 0.0946 | No | ||
| 80 | IKBKE | 67 | 14153 | -0.024 | 0.0963 | No | ||
| 81 | PIK3R3 | 5248 | 14193 | -0.025 | 0.0962 | No | ||
| 82 | MAP2K6 | 20614 1414 | 14483 | -0.030 | 0.0830 | No | ||
| 83 | FADD | 17536 8950 4711 | 14894 | -0.039 | 0.0640 | No | ||
| 84 | MYD88 | 18970 | 15863 | -0.073 | 0.0177 | No | ||
| 85 | STAT1 | 3936 5524 | 15868 | -0.073 | 0.0234 | No | ||
| 86 | CD80 | 22758 | 15922 | -0.076 | 0.0267 | No | ||
| 87 | MAPK8 | 6459 | 16496 | -0.113 | 0.0049 | No | ||
| 88 | JUN | 15832 | 16520 | -0.114 | 0.0129 | No | ||
| 89 | TLR2 | 15308 | 16615 | -0.123 | 0.0179 | No | ||
| 90 | MAP2K4 | 20405 | 16690 | -0.131 | 0.0245 | No | ||
| 91 | RAC1 | 16302 | 16844 | -0.147 | 0.0282 | No | ||
| 92 | PIK3CB | 19030 | 17205 | -0.186 | 0.0239 | No | ||
| 93 | TRAF6 | 5797 14940 | 17265 | -0.193 | 0.0364 | No | ||
| 94 | AKT3 | 13739 982 | 17357 | -0.204 | 0.0481 | No | ||
| 95 | MAPK1 | 1642 11167 | 17642 | -0.244 | 0.0526 | No |