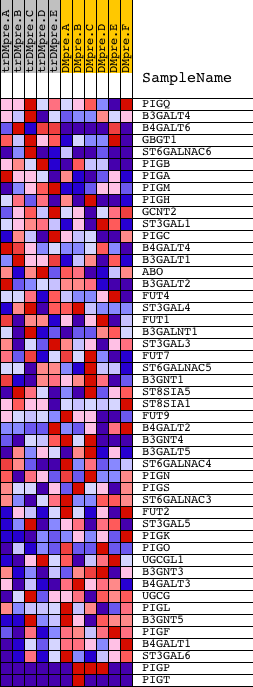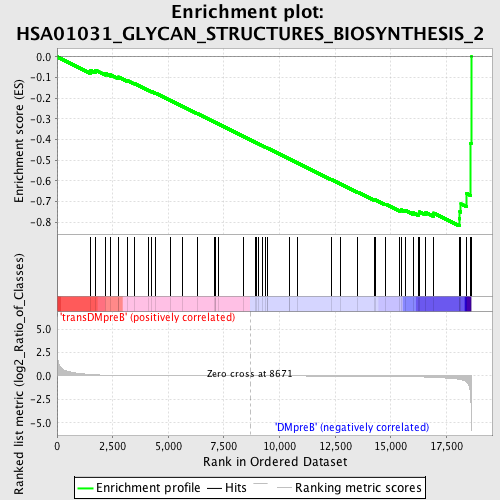
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | HSA01031_GLYCAN_STRUCTURES_BIOSYNTHESIS_2 |
| Enrichment Score (ES) | -0.8179199 |
| Normalized Enrichment Score (NES) | -1.751918 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022605738 |
| FWER p-Value | 0.032 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PIGQ | 4798 4797 | 1514 | 0.156 | -0.0657 | No | ||
| 2 | B3GALT4 | 23024 | 1732 | 0.125 | -0.0648 | No | ||
| 3 | B4GALT6 | 7109 12115 | 2159 | 0.078 | -0.0799 | No | ||
| 4 | GBGT1 | 15067 | 2397 | 0.062 | -0.0863 | No | ||
| 5 | ST6GALNAC6 | 11950 2856 | 2748 | 0.045 | -0.1007 | No | ||
| 6 | PIGB | 3128 12067 | 2766 | 0.044 | -0.0971 | No | ||
| 7 | PIGA | 2592 24210 | 3155 | 0.033 | -0.1147 | No | ||
| 8 | PIGM | 14046 | 3462 | 0.026 | -0.1286 | No | ||
| 9 | PIGH | 8483 | 4094 | 0.017 | -0.1609 | No | ||
| 10 | GCNT2 | 9168 3166 3243 21661 3270 | 4263 | 0.015 | -0.1684 | No | ||
| 11 | ST3GAL1 | 5435 2227 | 4425 | 0.014 | -0.1756 | No | ||
| 12 | PIGC | 14081 | 5116 | 0.010 | -0.2118 | No | ||
| 13 | B4GALT4 | 22756 | 5657 | 0.007 | -0.2401 | No | ||
| 14 | B3GALT1 | 6497 | 6319 | 0.005 | -0.2752 | No | ||
| 15 | ABO | 14647 | 7092 | 0.003 | -0.3165 | No | ||
| 16 | B3GALT2 | 6498 | 7136 | 0.003 | -0.3184 | No | ||
| 17 | FUT4 | 19240 | 7137 | 0.003 | -0.3181 | No | ||
| 18 | ST3GAL4 | 9816 | 7267 | 0.003 | -0.3248 | No | ||
| 19 | FUT1 | 8987 | 8399 | 0.001 | -0.3856 | No | ||
| 20 | B3GALNT1 | 15319 18224 | 8919 | -0.000 | -0.4135 | No | ||
| 21 | ST3GAL3 | 15782 | 8947 | -0.001 | -0.4149 | No | ||
| 22 | FUT7 | 15084 | 9035 | -0.001 | -0.4195 | No | ||
| 23 | ST6GALNAC5 | 15136 | 9228 | -0.001 | -0.4298 | No | ||
| 24 | B3GNT1 | 11997 | 9372 | -0.001 | -0.4373 | No | ||
| 25 | ST8SIA5 | 1950 1944 23508 | 9440 | -0.002 | -0.4408 | No | ||
| 26 | ST8SIA1 | 5437 | 9443 | -0.002 | -0.4407 | No | ||
| 27 | FUT9 | 16257 | 10440 | -0.004 | -0.4939 | No | ||
| 28 | B4GALT2 | 11990 | 10453 | -0.004 | -0.4942 | No | ||
| 29 | B3GNT4 | 10542 | 10816 | -0.005 | -0.5132 | No | ||
| 30 | B3GALT5 | 22690 | 12337 | -0.010 | -0.5940 | No | ||
| 31 | ST6GALNAC4 | 15036 2702 2848 2937 | 12749 | -0.012 | -0.6149 | No | ||
| 32 | PIGN | 13865 | 13505 | -0.018 | -0.6538 | No | ||
| 33 | PIGS | 11347 | 14243 | -0.026 | -0.6909 | No | ||
| 34 | ST6GALNAC3 | 2 | 14302 | -0.027 | -0.6914 | No | ||
| 35 | FUT2 | 17829 | 14753 | -0.035 | -0.7121 | No | ||
| 36 | ST3GAL5 | 17415 | 15394 | -0.052 | -0.7412 | No | ||
| 37 | PIGK | 1815 6834 11629 | 15475 | -0.055 | -0.7400 | No | ||
| 38 | PIGO | 15903 2488 2344 | 15652 | -0.063 | -0.7431 | No | ||
| 39 | UGCGL1 | 6721 | 16020 | -0.081 | -0.7547 | No | ||
| 40 | B3GNT3 | 18842 | 16265 | -0.097 | -0.7581 | No | ||
| 41 | B4GALT3 | 14057 | 16300 | -0.098 | -0.7500 | No | ||
| 42 | UGCG | 16197 | 16573 | -0.119 | -0.7526 | No | ||
| 43 | PIGL | 11595 | 16910 | -0.155 | -0.7551 | No | ||
| 44 | B3GNT5 | 22824 | 18077 | -0.348 | -0.7828 | Yes | ||
| 45 | PIGF | 22879 | 18104 | -0.357 | -0.7482 | Yes | ||
| 46 | B4GALT1 | 15918 | 18152 | -0.390 | -0.7114 | Yes | ||
| 47 | ST3GAL6 | 7046 | 18415 | -0.642 | -0.6608 | Yes | ||
| 48 | PIGP | 12082 1665 | 18593 | -2.484 | -0.4196 | Yes | ||
| 49 | PIGT | 8134 1 | 18611 | -4.171 | 0.0003 | Yes |