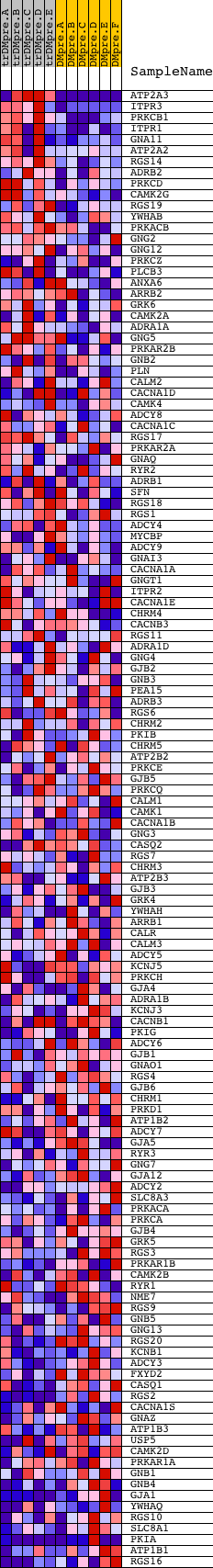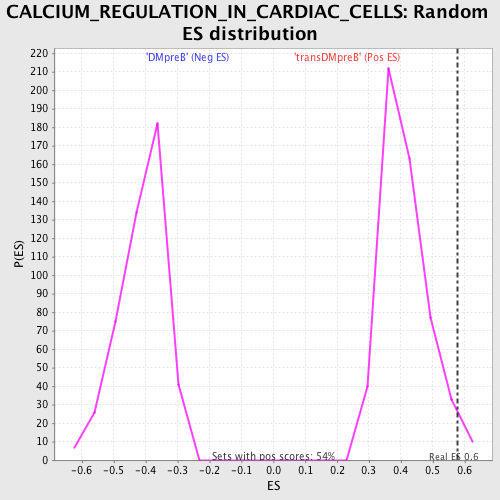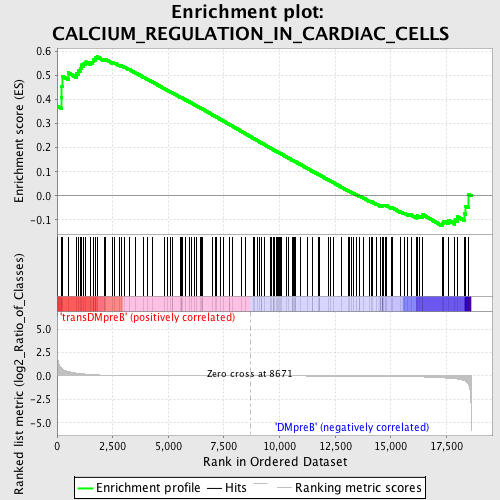
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | CALCIUM_REGULATION_IN_CARDIAC_CELLS |
| Enrichment Score (ES) | 0.5774157 |
| Normalized Enrichment Score (NES) | 1.4067342 |
| Nominal p-value | 0.028037382 |
| FDR q-value | 0.8999111 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 20795 1314 | 0 | 6.225 | 0.3724 | Yes | ||
| 2 | ITPR3 | 9195 | 202 | 0.792 | 0.4089 | Yes | ||
| 3 | PRKCB1 | 1693 9574 | 210 | 0.777 | 0.4550 | Yes | ||
| 4 | ITPR1 | 17341 | 237 | 0.732 | 0.4973 | Yes | ||
| 5 | GNA11 | 3300 3345 19670 | 505 | 0.472 | 0.5111 | Yes | ||
| 6 | ATP2A2 | 4421 3481 | 863 | 0.303 | 0.5100 | Yes | ||
| 7 | RGS14 | 21630 | 944 | 0.278 | 0.5223 | Yes | ||
| 8 | ADRB2 | 23422 | 1067 | 0.244 | 0.5303 | Yes | ||
| 9 | PRKCD | 21897 | 1081 | 0.237 | 0.5437 | Yes | ||
| 10 | CAMK2G | 21905 | 1204 | 0.210 | 0.5497 | Yes | ||
| 11 | RGS19 | 14303 | 1296 | 0.191 | 0.5562 | Yes | ||
| 12 | YWHAB | 14744 | 1492 | 0.160 | 0.5552 | Yes | ||
| 13 | PRKACB | 15140 | 1613 | 0.143 | 0.5573 | Yes | ||
| 14 | GNG2 | 4790 | 1631 | 0.139 | 0.5647 | Yes | ||
| 15 | GNG12 | 4789 | 1711 | 0.128 | 0.5681 | Yes | ||
| 16 | PRKCZ | 5260 | 1742 | 0.124 | 0.5739 | Yes | ||
| 17 | PLCB3 | 23799 | 1806 | 0.116 | 0.5774 | Yes | ||
| 18 | ANXA6 | 20449 | 2125 | 0.081 | 0.5651 | No | ||
| 19 | ARRB2 | 20806 | 2154 | 0.079 | 0.5683 | No | ||
| 20 | GRK6 | 6449 | 2484 | 0.057 | 0.5539 | No | ||
| 21 | CAMK2A | 2024 23541 1980 | 2597 | 0.051 | 0.5509 | No | ||
| 22 | ADRA1A | 2850 8560 | 2791 | 0.043 | 0.5430 | No | ||
| 23 | GNG5 | 9030 15393 483 | 2884 | 0.040 | 0.5405 | No | ||
| 24 | PRKAR2B | 5288 2107 | 3033 | 0.036 | 0.5346 | No | ||
| 25 | GNB2 | 9026 | 3247 | 0.030 | 0.5249 | No | ||
| 26 | PLN | 20026 | 3521 | 0.024 | 0.5116 | No | ||
| 27 | CALM2 | 8681 | 3889 | 0.019 | 0.4929 | No | ||
| 28 | CACNA1D | 4464 8675 | 4052 | 0.018 | 0.4852 | No | ||
| 29 | CAMK4 | 4473 | 4291 | 0.015 | 0.4732 | No | ||
| 30 | ADCY8 | 22281 | 4811 | 0.011 | 0.4458 | No | ||
| 31 | CACNA1C | 1158 1166 24464 4463 8674 | 4965 | 0.011 | 0.4381 | No | ||
| 32 | RGS17 | 7133 | 5103 | 0.010 | 0.4313 | No | ||
| 33 | PRKAR2A | 5287 | 5178 | 0.009 | 0.4279 | No | ||
| 34 | GNAQ | 4786 23909 3685 | 5192 | 0.009 | 0.4277 | No | ||
| 35 | RYR2 | 5403 | 5549 | 0.008 | 0.4089 | No | ||
| 36 | ADRB1 | 4357 | 5577 | 0.008 | 0.4079 | No | ||
| 37 | SFN | 7060 12062 | 5590 | 0.008 | 0.4077 | No | ||
| 38 | RGS18 | 13810 | 5622 | 0.008 | 0.4065 | No | ||
| 39 | RGS1 | 3970 6936 | 5783 | 0.007 | 0.3983 | No | ||
| 40 | ADCY4 | 2980 21818 | 5963 | 0.006 | 0.3890 | No | ||
| 41 | MYCBP | 7093 | 6049 | 0.006 | 0.3848 | No | ||
| 42 | ADCY9 | 22680 | 6176 | 0.006 | 0.3783 | No | ||
| 43 | GNAI3 | 15198 | 6268 | 0.006 | 0.3737 | No | ||
| 44 | CACNA1A | 4461 | 6462 | 0.005 | 0.3635 | No | ||
| 45 | GNGT1 | 17533 | 6483 | 0.005 | 0.3628 | No | ||
| 46 | ITPR2 | 9194 | 6512 | 0.005 | 0.3615 | No | ||
| 47 | CACNA1E | 4465 | 6521 | 0.005 | 0.3614 | No | ||
| 48 | CHRM4 | 14945 | 6998 | 0.004 | 0.3358 | No | ||
| 49 | CACNB3 | 22373 8679 | 7128 | 0.003 | 0.3291 | No | ||
| 50 | RGS11 | 23329 | 7169 | 0.003 | 0.3271 | No | ||
| 51 | ADRA1D | 20737 8561 | 7356 | 0.003 | 0.3172 | No | ||
| 52 | GNG4 | 21706 | 7493 | 0.002 | 0.3100 | No | ||
| 53 | GJB2 | 9018 21806 | 7743 | 0.002 | 0.2966 | No | ||
| 54 | GNB3 | 9027 | 7762 | 0.002 | 0.2957 | No | ||
| 55 | PEA15 | 13753 | 7887 | 0.002 | 0.2891 | No | ||
| 56 | ADRB3 | 18901 | 8301 | 0.001 | 0.2668 | No | ||
| 57 | RGS6 | 21219 | 8457 | 0.000 | 0.2585 | No | ||
| 58 | CHRM2 | 10840 | 8814 | -0.000 | 0.2392 | No | ||
| 59 | PKIB | 20022 | 8856 | -0.000 | 0.2370 | No | ||
| 60 | CHRM5 | 10038 | 8995 | -0.001 | 0.2296 | No | ||
| 61 | ATP2B2 | 17038 | 9111 | -0.001 | 0.2234 | No | ||
| 62 | PRKCE | 9575 | 9168 | -0.001 | 0.2205 | No | ||
| 63 | GJB5 | 15753 | 9189 | -0.001 | 0.2194 | No | ||
| 64 | PRKCQ | 2873 2831 | 9311 | -0.001 | 0.2130 | No | ||
| 65 | CALM1 | 21184 | 9605 | -0.002 | 0.1972 | No | ||
| 66 | CAMK1 | 1037 1170 17044 | 9641 | -0.002 | 0.1955 | No | ||
| 67 | CACNA1B | 8673 4462 | 9729 | -0.002 | 0.1909 | No | ||
| 68 | GNG3 | 23752 | 9732 | -0.002 | 0.1909 | No | ||
| 69 | CASQ2 | 15477 | 9785 | -0.002 | 0.1883 | No | ||
| 70 | RGS7 | 13743 | 9851 | -0.003 | 0.1849 | No | ||
| 71 | CHRM3 | 21547 | 9911 | -0.003 | 0.1819 | No | ||
| 72 | ATP2B3 | 24308 11557 | 9965 | -0.003 | 0.1792 | No | ||
| 73 | GJB3 | 15754 | 10016 | -0.003 | 0.1767 | No | ||
| 74 | GRK4 | 3467 16873 | 10040 | -0.003 | 0.1756 | No | ||
| 75 | YWHAH | 5937 10368 | 10098 | -0.003 | 0.1727 | No | ||
| 76 | ARRB1 | 3890 8465 18178 4276 | 10304 | -0.004 | 0.1618 | No | ||
| 77 | CALR | 18814 | 10389 | -0.004 | 0.1575 | No | ||
| 78 | CALM3 | 8682 | 10582 | -0.004 | 0.1474 | No | ||
| 79 | ADCY5 | 10312 | 10593 | -0.004 | 0.1471 | No | ||
| 80 | KCNJ5 | 9209 | 10632 | -0.004 | 0.1453 | No | ||
| 81 | PRKCH | 21246 | 10681 | -0.005 | 0.1430 | No | ||
| 82 | GJA4 | 15755 | 10694 | -0.005 | 0.1426 | No | ||
| 83 | ADRA1B | 4354 | 10736 | -0.005 | 0.1407 | No | ||
| 84 | KCNJ3 | 4944 | 10931 | -0.005 | 0.1305 | No | ||
| 85 | CACNB1 | 1443 1304 | 11269 | -0.006 | 0.1126 | No | ||
| 86 | PKIG | 9577 655 | 11477 | -0.007 | 0.1019 | No | ||
| 87 | ADCY6 | 22139 2283 8551 | 11485 | -0.007 | 0.1019 | No | ||
| 88 | GJB1 | 24276 | 11764 | -0.008 | 0.0873 | No | ||
| 89 | GNAO1 | 4785 3829 | 11779 | -0.008 | 0.0870 | No | ||
| 90 | RGS4 | 9724 | 11806 | -0.008 | 0.0861 | No | ||
| 91 | GJB6 | 21805 | 12205 | -0.009 | 0.0651 | No | ||
| 92 | CHRM1 | 23943 | 12308 | -0.010 | 0.0602 | No | ||
| 93 | PRKD1 | 2074 21075 | 12406 | -0.010 | 0.0556 | No | ||
| 94 | ATP1B2 | 20390 | 12798 | -0.013 | 0.0352 | No | ||
| 95 | ADCY7 | 8552 448 | 13095 | -0.014 | 0.0200 | No | ||
| 96 | GJA5 | 1929 9017 4779 | 13142 | -0.015 | 0.0184 | No | ||
| 97 | RYR3 | 14486 | 13238 | -0.015 | 0.0142 | No | ||
| 98 | GNG7 | 19684 | 13327 | -0.016 | 0.0104 | No | ||
| 99 | GJA12 | 20433 1323 | 13460 | -0.017 | 0.0042 | No | ||
| 100 | ADCY2 | 21412 | 13579 | -0.018 | -0.0010 | No | ||
| 101 | SLC8A3 | 2065 4293 | 13601 | -0.018 | -0.0011 | No | ||
| 102 | PRKACA | 18549 3844 | 13785 | -0.020 | -0.0098 | No | ||
| 103 | PRKCA | 20174 | 14043 | -0.023 | -0.0223 | No | ||
| 104 | GJB4 | 4782 | 14137 | -0.024 | -0.0259 | No | ||
| 105 | GRK5 | 9036 23814 | 14155 | -0.024 | -0.0254 | No | ||
| 106 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 14351 | -0.027 | -0.0343 | No | ||
| 107 | PRKAR1B | 16320 | 14524 | -0.031 | -0.0417 | No | ||
| 108 | CAMK2B | 20536 | 14553 | -0.031 | -0.0414 | No | ||
| 109 | RYR1 | 17903 17902 9770 | 14603 | -0.032 | -0.0421 | No | ||
| 110 | NME7 | 926 4101 14070 3986 3946 931 | 14626 | -0.033 | -0.0414 | No | ||
| 111 | RGS9 | 20172 | 14628 | -0.033 | -0.0395 | No | ||
| 112 | GNB5 | 2985 9029 3153 | 14647 | -0.033 | -0.0385 | No | ||
| 113 | GNG13 | 23338 | 14689 | -0.034 | -0.0387 | No | ||
| 114 | RGS20 | 4001 12202 | 14744 | -0.035 | -0.0395 | No | ||
| 115 | KCNB1 | 14334 | 14818 | -0.037 | -0.0412 | No | ||
| 116 | ADCY3 | 21329 894 | 15046 | -0.042 | -0.0510 | No | ||
| 117 | FXYD2 | 3038 19470 3068 3039 | 15058 | -0.042 | -0.0491 | No | ||
| 118 | CASQ1 | 13752 8695 | 15416 | -0.053 | -0.0652 | No | ||
| 119 | RGS2 | 5379 | 15602 | -0.061 | -0.0715 | No | ||
| 120 | CACNA1S | 14111 | 15766 | -0.068 | -0.0763 | No | ||
| 121 | GNAZ | 71 | 15908 | -0.075 | -0.0794 | No | ||
| 122 | ATP1B3 | 19037 | 16149 | -0.089 | -0.0871 | No | ||
| 123 | USP5 | 17004 | 16177 | -0.091 | -0.0831 | No | ||
| 124 | CAMK2D | 4232 | 16308 | -0.099 | -0.0842 | No | ||
| 125 | PRKAR1A | 20615 | 16423 | -0.108 | -0.0840 | No | ||
| 126 | GNB1 | 15967 | 16431 | -0.109 | -0.0778 | No | ||
| 127 | GNB4 | 4787 9028 | 17311 | -0.199 | -0.1135 | No | ||
| 128 | GJA1 | 20023 | 17380 | -0.207 | -0.1048 | No | ||
| 129 | YWHAQ | 10369 | 17583 | -0.236 | -0.1016 | No | ||
| 130 | RGS10 | 12591 | 17883 | -0.295 | -0.1001 | No | ||
| 131 | SLC8A1 | 9830 | 17998 | -0.323 | -0.0870 | No | ||
| 132 | PKIA | 15639 | 18316 | -0.497 | -0.0744 | No | ||
| 133 | ATP1B1 | 4420 | 18372 | -0.566 | -0.0435 | No | ||
| 134 | RGS16 | 14094 | 18496 | -0.946 | 0.0065 | No |