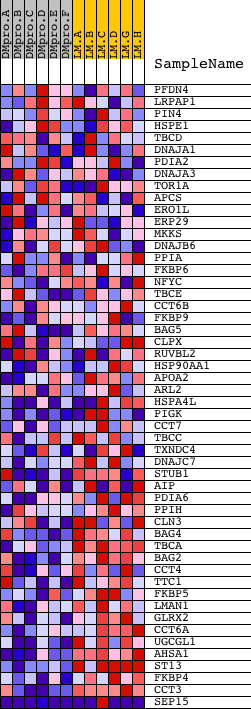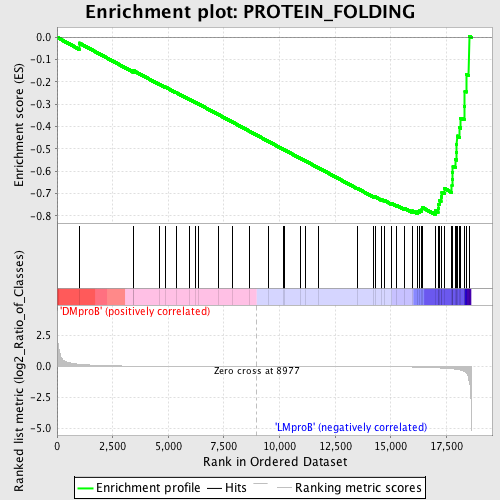
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | PROTEIN_FOLDING |
| Enrichment Score (ES) | -0.79426384 |
| Normalized Enrichment Score (NES) | -1.6380335 |
| Nominal p-value | 0.0021551724 |
| FDR q-value | 0.6277486 |
| FWER p-Value | 0.68 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PFDN4 | 4260 | 992 | 0.176 | -0.0253 | No | ||
| 2 | LRPAP1 | 5005 | 3434 | 0.023 | -0.1530 | No | ||
| 3 | PIN4 | 12817 | 3441 | 0.023 | -0.1497 | No | ||
| 4 | HSPE1 | 9133 | 4607 | 0.012 | -0.2105 | No | ||
| 5 | TBCD | 20555 | 4861 | 0.010 | -0.2225 | No | ||
| 6 | DNAJA1 | 4878 16242 2345 | 5352 | 0.008 | -0.2476 | No | ||
| 7 | PDIA2 | 23062 22604 1702 | 5942 | 0.006 | -0.2783 | No | ||
| 8 | DNAJA3 | 1732 13518 | 6220 | 0.006 | -0.2923 | No | ||
| 9 | TOR1A | 11395 2909 | 6333 | 0.005 | -0.2975 | No | ||
| 10 | APCS | 13748 | 7265 | 0.003 | -0.3471 | No | ||
| 11 | ERO1L | 6930 11924 6929 | 7881 | 0.002 | -0.3799 | No | ||
| 12 | ERP29 | 12524 | 8665 | 0.001 | -0.4220 | No | ||
| 13 | MKKS | 2946 14416 | 9504 | -0.001 | -0.4670 | No | ||
| 14 | DNAJB6 | 6246 3538 | 10186 | -0.002 | -0.5034 | No | ||
| 15 | PPIA | 1284 11188 | 10226 | -0.002 | -0.5052 | No | ||
| 16 | FKBP6 | 16344 | 10958 | -0.003 | -0.5440 | No | ||
| 17 | NFYC | 9460 | 11178 | -0.004 | -0.5552 | No | ||
| 18 | TBCE | 21543 | 11740 | -0.005 | -0.5846 | No | ||
| 19 | CCT6B | 20326 | 13490 | -0.011 | -0.6771 | No | ||
| 20 | FKBP9 | 17433 | 14230 | -0.015 | -0.7145 | No | ||
| 21 | BAG5 | 20981 192 | 14289 | -0.016 | -0.7151 | No | ||
| 22 | CLPX | 19403 | 14597 | -0.018 | -0.7287 | No | ||
| 23 | RUVBL2 | 9766 | 14696 | -0.019 | -0.7309 | No | ||
| 24 | HSP90AA1 | 4883 4882 9131 | 15009 | -0.023 | -0.7440 | No | ||
| 25 | APOA2 | 8615 4044 | 15265 | -0.027 | -0.7534 | No | ||
| 26 | ARL2 | 23992 | 15605 | -0.035 | -0.7662 | No | ||
| 27 | HSPA4L | 5214 | 15967 | -0.045 | -0.7783 | No | ||
| 28 | PIGK | 1815 6834 11629 | 16212 | -0.056 | -0.7825 | Yes | ||
| 29 | CCT7 | 17385 | 16300 | -0.060 | -0.7776 | Yes | ||
| 30 | TBCC | 23215 | 16396 | -0.064 | -0.7724 | Yes | ||
| 31 | TXNDC4 | 15886 | 16402 | -0.065 | -0.7623 | Yes | ||
| 32 | DNAJC7 | 20227 | 16996 | -0.104 | -0.7776 | Yes | ||
| 33 | STUB1 | 23070 | 17123 | -0.115 | -0.7661 | Yes | ||
| 34 | AIP | 23955 | 17151 | -0.117 | -0.7488 | Yes | ||
| 35 | PDIA6 | 21308 | 17197 | -0.122 | -0.7317 | Yes | ||
| 36 | PPIH | 12287 | 17258 | -0.129 | -0.7142 | Yes | ||
| 37 | CLN3 | 17635 | 17296 | -0.133 | -0.6949 | Yes | ||
| 38 | BAG4 | 7431 | 17407 | -0.144 | -0.6778 | Yes | ||
| 39 | TBCA | 10037 | 17747 | -0.187 | -0.6661 | Yes | ||
| 40 | BAG2 | 13985 | 17753 | -0.188 | -0.6364 | Yes | ||
| 41 | CCT4 | 8710 | 17774 | -0.191 | -0.6069 | Yes | ||
| 42 | TTC1 | 7342 | 17793 | -0.194 | -0.5769 | Yes | ||
| 43 | FKBP5 | 23051 | 17911 | -0.216 | -0.5487 | Yes | ||
| 44 | LMAN1 | 12867 | 17942 | -0.224 | -0.5145 | Yes | ||
| 45 | GLRX2 | 12795 | 17964 | -0.230 | -0.4788 | Yes | ||
| 46 | CCT6A | 16689 | 17974 | -0.232 | -0.4422 | Yes | ||
| 47 | UGCGL1 | 6721 | 18095 | -0.272 | -0.4052 | Yes | ||
| 48 | AHSA1 | 21192 | 18143 | -0.287 | -0.3618 | Yes | ||
| 49 | ST13 | 12865 | 18297 | -0.389 | -0.3078 | Yes | ||
| 50 | FKBP4 | 4726 | 18332 | -0.421 | -0.2422 | Yes | ||
| 51 | CCT3 | 8709 | 18387 | -0.494 | -0.1660 | Yes | ||
| 52 | SEP15 | 13527 1895 15398 | 18513 | -1.114 | 0.0055 | Yes |