Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | WGGAATGY_V$TEF1_Q6 |
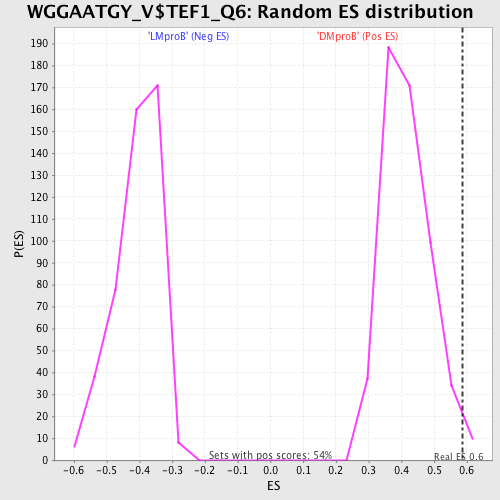
| Enrichment Score (ES) | 0.58563304 |
| Normalized Enrichment Score (NES) | 1.4065548 |
| Nominal p-value | 0.018552875 |
| FDR q-value | 0.8252167 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 3 | 4.234 | 0.2116 | Yes | ||
| 2 | RASSF2 | 2802 14422 | 120 | 1.156 | 0.2632 | Yes | ||
| 3 | RHBDF1 | 20504 | 138 | 1.004 | 0.3125 | Yes | ||
| 4 | ITGB1 | 3872 18411 | 188 | 0.692 | 0.3445 | Yes | ||
| 5 | ARHGAP24 | 16778 3655 | 233 | 0.570 | 0.3706 | Yes | ||
| 6 | BCL11A | 4691 | 318 | 0.446 | 0.3883 | Yes | ||
| 7 | NFKB2 | 23810 | 323 | 0.440 | 0.4101 | Yes | ||
| 8 | ITPR3 | 9195 | 334 | 0.424 | 0.4307 | Yes | ||
| 9 | SH3BGRL3 | 15722 | 481 | 0.322 | 0.4389 | Yes | ||
| 10 | SIPA1 | 9820 | 604 | 0.270 | 0.4458 | Yes | ||
| 11 | RASGRP2 | 9693 | 611 | 0.269 | 0.4589 | Yes | ||
| 12 | FRAG1 | 6129 18165 1724 | 690 | 0.242 | 0.4668 | Yes | ||
| 13 | VCL | 22083 | 701 | 0.238 | 0.4781 | Yes | ||
| 14 | HMGA1 | 23323 | 705 | 0.236 | 0.4898 | Yes | ||
| 15 | TNNT2 | 14113 | 796 | 0.212 | 0.4955 | Yes | ||
| 16 | MBNL2 | 21932 | 859 | 0.199 | 0.5021 | Yes | ||
| 17 | RRAS | 18256 | 891 | 0.194 | 0.5101 | Yes | ||
| 18 | SMAD3 | 19084 | 925 | 0.187 | 0.5177 | Yes | ||
| 19 | CDC42EP1 | 22433 10435 13411 11239 | 927 | 0.187 | 0.5270 | Yes | ||
| 20 | DDIT3 | 19857 | 1073 | 0.163 | 0.5272 | Yes | ||
| 21 | PHF12 | 11192 6483 | 1103 | 0.158 | 0.5336 | Yes | ||
| 22 | PGF | 21020 | 1215 | 0.143 | 0.5347 | Yes | ||
| 23 | MYADM | 11946 | 1261 | 0.137 | 0.5391 | Yes | ||
| 24 | GOLGA1 | 14595 | 1271 | 0.136 | 0.5455 | Yes | ||
| 25 | LAMB3 | 14007 | 1292 | 0.133 | 0.5510 | Yes | ||
| 26 | ST5 | 17682 | 1408 | 0.120 | 0.5508 | Yes | ||
| 27 | HDGF | 9082 4846 4845 | 1488 | 0.112 | 0.5521 | Yes | ||
| 28 | IL6ST | 4920 | 1572 | 0.104 | 0.5527 | Yes | ||
| 29 | APOE | 8618 | 1645 | 0.098 | 0.5537 | Yes | ||
| 30 | TRAF4 | 10217 5796 1400 | 1656 | 0.097 | 0.5580 | Yes | ||
| 31 | EHD1 | 23791 | 1662 | 0.096 | 0.5625 | Yes | ||
| 32 | FGFR2 | 1917 4722 1119 | 1673 | 0.095 | 0.5667 | Yes | ||
| 33 | DMD | 24295 2647 | 1698 | 0.093 | 0.5701 | Yes | ||
| 34 | SCNN1A | 17278 | 1710 | 0.092 | 0.5741 | Yes | ||
| 35 | BMP4 | 21863 | 1731 | 0.091 | 0.5775 | Yes | ||
| 36 | CDK2 | 3438 3373 19592 3322 | 1735 | 0.090 | 0.5819 | Yes | ||
| 37 | CNN3 | 12983 | 1749 | 0.089 | 0.5856 | Yes | ||
| 38 | SLC26A6 | 19304 | 2245 | 0.058 | 0.5616 | No | ||
| 39 | SIN3A | 5442 | 2277 | 0.057 | 0.5628 | No | ||
| 40 | ATP5G2 | 12610 | 2308 | 0.056 | 0.5639 | No | ||
| 41 | ITGB4 | 9669 159 | 2592 | 0.044 | 0.5507 | No | ||
| 42 | MID1 | 5097 5098 | 2759 | 0.038 | 0.5436 | No | ||
| 43 | FGF13 | 4720 8963 2627 | 2794 | 0.037 | 0.5435 | No | ||
| 44 | STARD3 | 20675 | 2919 | 0.033 | 0.5385 | No | ||
| 45 | FSTL1 | 22765 | 2937 | 0.033 | 0.5392 | No | ||
| 46 | MYH7 | 21828 4709 | 2950 | 0.033 | 0.5402 | No | ||
| 47 | DDX3X | 24379 | 2969 | 0.032 | 0.5408 | No | ||
| 48 | IL17RE | 434 17327 433 1049 | 3264 | 0.026 | 0.5261 | No | ||
| 49 | APPBP2 | 7357 | 3364 | 0.024 | 0.5219 | No | ||
| 50 | DUSP7 | 10661 | 3370 | 0.024 | 0.5229 | No | ||
| 51 | HAPLN1 | 21592 | 3488 | 0.022 | 0.5176 | No | ||
| 52 | PKP4 | 2822 15008 | 3546 | 0.022 | 0.5156 | No | ||
| 53 | ADAM33 | 14426 | 3569 | 0.021 | 0.5155 | No | ||
| 54 | CSRP2BP | 2696 10458 | 3698 | 0.019 | 0.5095 | No | ||
| 55 | HOOK1 | 2395 13418 8102 16173 | 3700 | 0.019 | 0.5104 | No | ||
| 56 | SYT8 | 18006 | 3757 | 0.019 | 0.5083 | No | ||
| 57 | SPRED1 | 4331 | 3849 | 0.018 | 0.5042 | No | ||
| 58 | TGIF2 | 6013 6012 | 3881 | 0.017 | 0.5034 | No | ||
| 59 | POPDC2 | 22760 | 3916 | 0.017 | 0.5024 | No | ||
| 60 | MNT | 20784 | 3972 | 0.016 | 0.5002 | No | ||
| 61 | NOG | 13663 | 4054 | 0.016 | 0.4966 | No | ||
| 62 | MAPK10 | 11169 | 4304 | 0.014 | 0.4837 | No | ||
| 63 | SH3BP1 | 632 22431 | 4318 | 0.013 | 0.4837 | No | ||
| 64 | GREM1 | 14485 | 4322 | 0.013 | 0.4842 | No | ||
| 65 | COL5A3 | 19219 | 4329 | 0.013 | 0.4845 | No | ||
| 66 | MLLT3 | 2359 15847 2413 | 4544 | 0.012 | 0.4735 | No | ||
| 67 | HOXB3 | 20685 | 4603 | 0.012 | 0.4709 | No | ||
| 68 | RBMS3 | 18973 | 4619 | 0.012 | 0.4707 | No | ||
| 69 | SOSTDC1 | 21286 | 4692 | 0.011 | 0.4673 | No | ||
| 70 | CHRNA1 | 4319 14559 8530 | 4743 | 0.011 | 0.4652 | No | ||
| 71 | KIFC3 | 9227 | 4771 | 0.011 | 0.4642 | No | ||
| 72 | BMPR2 | 14241 4081 | 4816 | 0.011 | 0.4624 | No | ||
| 73 | WDR13 | 7859 13112 | 4856 | 0.010 | 0.4608 | No | ||
| 74 | JUB | 21834 3339 | 4903 | 0.010 | 0.4588 | No | ||
| 75 | RPH3AL | 11725 | 4941 | 0.010 | 0.4573 | No | ||
| 76 | ZIC1 | 19040 | 4985 | 0.010 | 0.4554 | No | ||
| 77 | ESR1 | 20097 4685 | 5004 | 0.010 | 0.4549 | No | ||
| 78 | PIAS3 | 15491 672 1906 | 5078 | 0.009 | 0.4514 | No | ||
| 79 | TIAL1 | 996 5753 | 5079 | 0.009 | 0.4519 | No | ||
| 80 | NAP1L5 | 17134 | 5210 | 0.009 | 0.4453 | No | ||
| 81 | PRX | 3729 9628 | 5294 | 0.009 | 0.4412 | No | ||
| 82 | HOXA10 | 17145 989 | 5413 | 0.008 | 0.4352 | No | ||
| 83 | SPARC | 20444 | 5422 | 0.008 | 0.4352 | No | ||
| 84 | NSD1 | 2134 5197 | 5424 | 0.008 | 0.4355 | No | ||
| 85 | COL11A2 | 23286 | 5536 | 0.008 | 0.4298 | No | ||
| 86 | FBXL13 | 16596 | 5562 | 0.008 | 0.4289 | No | ||
| 87 | CACNA1G | 1237 940 20292 | 5717 | 0.007 | 0.4208 | No | ||
| 88 | OTX2 | 9519 | 5727 | 0.007 | 0.4207 | No | ||
| 89 | ITGA3 | 20284 | 5977 | 0.006 | 0.4075 | No | ||
| 90 | SMOC2 | 23378 | 6095 | 0.006 | 0.4014 | No | ||
| 91 | BMP5 | 19376 | 6111 | 0.006 | 0.4009 | No | ||
| 92 | SLC12A4 | 18763 | 6194 | 0.006 | 0.3967 | No | ||
| 93 | LIFR | 22515 9277 | 6294 | 0.005 | 0.3916 | No | ||
| 94 | NR2E1 | 19780 | 6449 | 0.005 | 0.3834 | No | ||
| 95 | IRS4 | 9183 4926 | 6467 | 0.005 | 0.3828 | No | ||
| 96 | MUSK | 5199 | 6607 | 0.005 | 0.3754 | No | ||
| 97 | LRRC1 | 19059 3129 | 6611 | 0.005 | 0.3755 | No | ||
| 98 | CFL2 | 2155 21069 | 6622 | 0.005 | 0.3752 | No | ||
| 99 | VIT | 23155 | 6673 | 0.004 | 0.3727 | No | ||
| 100 | REST | 16818 | 6743 | 0.004 | 0.3691 | No | ||
| 101 | COL4A6 | 8248 2552 | 6905 | 0.004 | 0.3606 | No | ||
| 102 | ZNF462 | 6321 | 6930 | 0.004 | 0.3594 | No | ||
| 103 | PTPRJ | 9664 | 7224 | 0.003 | 0.3437 | No | ||
| 104 | SLC9A3R1 | 11250 | 7314 | 0.003 | 0.3390 | No | ||
| 105 | CRLF1 | 18588 | 7421 | 0.003 | 0.3333 | No | ||
| 106 | OLFML3 | 15216 | 7477 | 0.003 | 0.3305 | No | ||
| 107 | GLP2R | 8227 13563 13564 | 7657 | 0.002 | 0.3209 | No | ||
| 108 | SYT2 | 14122 | 7706 | 0.002 | 0.3184 | No | ||
| 109 | NEDD4 | 19384 3101 | 7900 | 0.002 | 0.3080 | No | ||
| 110 | KLK6 | 9231 9626 | 7914 | 0.002 | 0.3073 | No | ||
| 111 | MPZ | 9408 | 7924 | 0.002 | 0.3070 | No | ||
| 112 | UPK1B | 22592 | 7960 | 0.002 | 0.3051 | No | ||
| 113 | PDGFRA | 16824 | 8042 | 0.002 | 0.3008 | No | ||
| 114 | SILV | 16130 | 8130 | 0.001 | 0.2962 | No | ||
| 115 | CLDN2 | 4527 | 8178 | 0.001 | 0.2937 | No | ||
| 116 | NR4A3 | 9473 16212 5183 | 8351 | 0.001 | 0.2844 | No | ||
| 117 | BMP10 | 17376 | 8365 | 0.001 | 0.2837 | No | ||
| 118 | PRDM1 | 19775 3337 | 8528 | 0.001 | 0.2749 | No | ||
| 119 | PROK2 | 17057 | 8557 | 0.001 | 0.2734 | No | ||
| 120 | PDGFA | 5237 | 8589 | 0.001 | 0.2718 | No | ||
| 121 | POGK | 7739 | 8639 | 0.001 | 0.2691 | No | ||
| 122 | LHX4 | 13800 | 9173 | -0.000 | 0.2401 | No | ||
| 123 | JMJD1C | 11531 19996 | 9199 | -0.000 | 0.2388 | No | ||
| 124 | SCARF2 | 22833 | 9230 | -0.000 | 0.2372 | No | ||
| 125 | KCNH2 | 16592 | 9241 | -0.000 | 0.2367 | No | ||
| 126 | MTSS1 | 5585 22285 | 9627 | -0.001 | 0.2158 | No | ||
| 127 | SLC12A5 | 14731 | 9639 | -0.001 | 0.2152 | No | ||
| 128 | NOTCH2 | 15485 | 9669 | -0.001 | 0.2137 | No | ||
| 129 | GPR21 | 15020 | 9683 | -0.001 | 0.2131 | No | ||
| 130 | EFNB3 | 20391 | 9703 | -0.001 | 0.2121 | No | ||
| 131 | PRKCI | 9576 | 9792 | -0.001 | 0.2074 | No | ||
| 132 | EDN1 | 21658 | 9812 | -0.001 | 0.2064 | No | ||
| 133 | CTGF | 20068 | 9870 | -0.001 | 0.2034 | No | ||
| 134 | PLSCR2 | 19354 | 9902 | -0.001 | 0.2017 | No | ||
| 135 | NNMT | 19131 | 9961 | -0.002 | 0.1987 | No | ||
| 136 | DVL3 | 8873 22821 | 9986 | -0.002 | 0.1974 | No | ||
| 137 | PTRF | 9667 | 10002 | -0.002 | 0.1967 | No | ||
| 138 | NRG1 | 3885 | 10016 | -0.002 | 0.1961 | No | ||
| 139 | ACSL3 | 13184 | 10040 | -0.002 | 0.1949 | No | ||
| 140 | KIF1C | 4951 | 10054 | -0.002 | 0.1943 | No | ||
| 141 | HMGB2 | 13594 | 10162 | -0.002 | 0.1886 | No | ||
| 142 | COL4A5 | 4543 | 10174 | -0.002 | 0.1881 | No | ||
| 143 | PGAM2 | 20539 | 10393 | -0.002 | 0.1763 | No | ||
| 144 | NR4A1 | 9099 | 10437 | -0.002 | 0.1741 | No | ||
| 145 | MAPRE3 | 8301 | 10489 | -0.003 | 0.1715 | No | ||
| 146 | ADPRHL1 | 18930 3863 | 10493 | -0.003 | 0.1714 | No | ||
| 147 | PTPN11 | 5326 16391 9660 | 10593 | -0.003 | 0.1662 | No | ||
| 148 | IER5 | 13802 | 11026 | -0.004 | 0.1428 | No | ||
| 149 | CGN | 12896 12897 7701 | 11194 | -0.004 | 0.1339 | No | ||
| 150 | NES | 5162 9454 | 11273 | -0.004 | 0.1299 | No | ||
| 151 | GPM6A | 3834 10618 | 11349 | -0.004 | 0.1260 | No | ||
| 152 | PAQR6 | 15551 12756 7591 | 11384 | -0.004 | 0.1244 | No | ||
| 153 | SIX4 | 5445 | 11445 | -0.004 | 0.1213 | No | ||
| 154 | SOST | 20210 | 11507 | -0.005 | 0.1182 | No | ||
| 155 | CDH3 | 3919 4509 | 11519 | -0.005 | 0.1179 | No | ||
| 156 | ITGB1BP2 | 24275 | 11539 | -0.005 | 0.1171 | No | ||
| 157 | LMOD1 | 14119 | 11770 | -0.005 | 0.1048 | No | ||
| 158 | MYH6 | 9436 | 11865 | -0.005 | 0.1000 | No | ||
| 159 | NDST4 | 12259 7228 | 11939 | -0.006 | 0.0963 | No | ||
| 160 | DACT1 | 7203 | 11978 | -0.006 | 0.0945 | No | ||
| 161 | TRPC4 | 15593 | 12011 | -0.006 | 0.0930 | No | ||
| 162 | MCF2 | 24147 | 12203 | -0.006 | 0.0829 | No | ||
| 163 | PROP1 | 20472 | 12215 | -0.006 | 0.0827 | No | ||
| 164 | NRG2 | 4274 | 12362 | -0.007 | 0.0750 | No | ||
| 165 | FOXN1 | 20337 | 12400 | -0.007 | 0.0734 | No | ||
| 166 | RAPSN | 2879 14951 | 12507 | -0.007 | 0.0679 | No | ||
| 167 | HAND2 | 18615 | 12592 | -0.007 | 0.0637 | No | ||
| 168 | RARG | 22113 | 12596 | -0.007 | 0.0639 | No | ||
| 169 | GHRL | 1183 17040 | 12733 | -0.008 | 0.0569 | No | ||
| 170 | KLHDC3 | 22956 | 12880 | -0.008 | 0.0494 | No | ||
| 171 | T | 23388 | 13002 | -0.009 | 0.0432 | No | ||
| 172 | LIN28 | 15723 | 13032 | -0.009 | 0.0421 | No | ||
| 173 | HOXC13 | 22344 | 13107 | -0.009 | 0.0385 | No | ||
| 174 | CAV1 | 8698 | 13216 | -0.009 | 0.0331 | No | ||
| 175 | MSX2 | 21461 | 13217 | -0.009 | 0.0335 | No | ||
| 176 | MRGPRF | 17995 | 13283 | -0.010 | 0.0305 | No | ||
| 177 | CPNE1 | 11186 | 13297 | -0.010 | 0.0303 | No | ||
| 178 | CLDN16 | 22802 | 13314 | -0.010 | 0.0299 | No | ||
| 179 | ERG | 1686 8915 | 13370 | -0.010 | 0.0274 | No | ||
| 180 | ATRN | 4432 2964 | 13448 | -0.010 | 0.0237 | No | ||
| 181 | PLXNB1 | 19301 | 13572 | -0.011 | 0.0176 | No | ||
| 182 | ERBB2 | 8913 | 13586 | -0.011 | 0.0174 | No | ||
| 183 | PDE1A | 9541 5234 | 13627 | -0.011 | 0.0158 | No | ||
| 184 | GPRC5C | 12864 | 13655 | -0.011 | 0.0149 | No | ||
| 185 | SHC1 | 9813 9812 5430 | 13709 | -0.012 | 0.0126 | No | ||
| 186 | ACTA1 | 18715 | 13818 | -0.013 | 0.0074 | No | ||
| 187 | TRPM3 | 23904 10357 5927 | 13836 | -0.013 | 0.0071 | No | ||
| 188 | PRKCDBP | 17706 | 13954 | -0.013 | 0.0014 | No | ||
| 189 | FGA | 1780 15566 | 14005 | -0.014 | -0.0007 | No | ||
| 190 | LAMC2 | 4983 9266 13806 3975 | 14094 | -0.014 | -0.0047 | No | ||
| 191 | SYTL2 | 18190 1539 3730 | 14170 | -0.015 | -0.0081 | No | ||
| 192 | DDR1 | 1625 23000 | 14206 | -0.015 | -0.0092 | No | ||
| 193 | GABRB1 | 16832 | 14217 | -0.015 | -0.0090 | No | ||
| 194 | SCN8A | 5414 | 14260 | -0.015 | -0.0105 | No | ||
| 195 | HSPB2 | 19121 | 14278 | -0.016 | -0.0107 | No | ||
| 196 | IGFBP6 | 9161 | 14287 | -0.016 | -0.0103 | No | ||
| 197 | FOXP2 | 4312 17523 8523 | 14306 | -0.016 | -0.0105 | No | ||
| 198 | MGLL | 17368 1145 | 14312 | -0.016 | -0.0100 | No | ||
| 199 | ELF3 | 8894 13824 | 14464 | -0.017 | -0.0174 | No | ||
| 200 | MAN1A2 | 5076 5075 | 14486 | -0.017 | -0.0177 | No | ||
| 201 | AVPI1 | 23675 | 14491 | -0.017 | -0.0170 | No | ||
| 202 | CNTN6 | 17346 | 14503 | -0.017 | -0.0167 | No | ||
| 203 | SPDEF | 23055 | 14561 | -0.018 | -0.0189 | No | ||
| 204 | FGFR4 | 3226 | 14823 | -0.021 | -0.0321 | No | ||
| 205 | SLC5A2 | 18057 10905 | 14901 | -0.022 | -0.0352 | No | ||
| 206 | POGZ | 6031 1911 | 14913 | -0.022 | -0.0347 | No | ||
| 207 | TMSB10 | 5322 | 14995 | -0.023 | -0.0380 | No | ||
| 208 | NTF5 | 18248 | 15031 | -0.023 | -0.0388 | No | ||
| 209 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 15048 | -0.024 | -0.0384 | No | ||
| 210 | PPIL4 | 20092 | 15070 | -0.024 | -0.0384 | No | ||
| 211 | ANGPT1 | 8588 2260 22303 | 15086 | -0.024 | -0.0380 | No | ||
| 212 | PTHR1 | 5318 18985 | 15137 | -0.025 | -0.0395 | No | ||
| 213 | RAB11B | 9676 | 15178 | -0.026 | -0.0404 | No | ||
| 214 | SLC16A8 | 22210 | 15215 | -0.026 | -0.0410 | No | ||
| 215 | MAST2 | 9423 | 15310 | -0.028 | -0.0448 | No | ||
| 216 | MDK | 14523 | 15523 | -0.033 | -0.0547 | No | ||
| 217 | PPFIA1 | 17538 6136 | 15555 | -0.033 | -0.0547 | No | ||
| 218 | ROCK2 | 21309 5387 | 15591 | -0.034 | -0.0549 | No | ||
| 219 | CHIC2 | 16510 | 15657 | -0.036 | -0.0566 | No | ||
| 220 | KCNE2 | 22701 | 15801 | -0.040 | -0.0624 | No | ||
| 221 | CYR61 | 9159 15145 9311 5024 | 15858 | -0.042 | -0.0634 | No | ||
| 222 | RNF2 | 9728 | 15900 | -0.043 | -0.0634 | No | ||
| 223 | LMNA | 4999 15290 1778 9280 | 16070 | -0.050 | -0.0701 | No | ||
| 224 | TENC1 | 22349 9927 | 16132 | -0.052 | -0.0709 | No | ||
| 225 | PCBP1 | 17085 | 16486 | -0.068 | -0.0866 | No | ||
| 226 | AP2M1 | 8603 | 16556 | -0.072 | -0.0868 | No | ||
| 227 | CDK6 | 16600 | 16668 | -0.079 | -0.0889 | No | ||
| 228 | LAMB2 | 9265 | 16687 | -0.081 | -0.0858 | No | ||
| 229 | SH3BGRL2 | 5601 | 16891 | -0.096 | -0.0921 | No | ||
| 230 | PLS3 | 8332 | 16979 | -0.102 | -0.0917 | No | ||
| 231 | BTF3 | 10176 | 17022 | -0.106 | -0.0887 | No | ||
| 232 | FHOD1 | 18772 | 17047 | -0.108 | -0.0846 | No | ||
| 233 | TPM1 | 19070 | 17146 | -0.117 | -0.0841 | No | ||
| 234 | RNF19 | 22314 | 17257 | -0.129 | -0.0836 | No | ||
| 235 | GALNT12 | 16214 | 17691 | -0.179 | -0.0982 | No | ||
| 236 | TMOD3 | 6945 | 17714 | -0.182 | -0.0903 | No | ||
| 237 | PIK3C3 | 23607 | 17759 | -0.189 | -0.0833 | No | ||
| 238 | EPHA2 | 16006 | 18060 | -0.262 | -0.0865 | No | ||
| 239 | CRYAB | 19457 | 18139 | -0.286 | -0.0764 | No | ||
| 240 | SLC31A2 | 9827 | 18144 | -0.288 | -0.0622 | No | ||
| 241 | CHD4 | 8418 17281 4225 | 18163 | -0.297 | -0.0484 | No | ||
| 242 | ATF4 | 22417 2239 | 18251 | -0.354 | -0.0354 | No | ||
| 243 | CDKN2C | 15806 2321 | 18342 | -0.432 | -0.0187 | No | ||
| 244 | GDI1 | 9012 4770 24297 4769 | 18448 | -0.671 | 0.0091 | No |