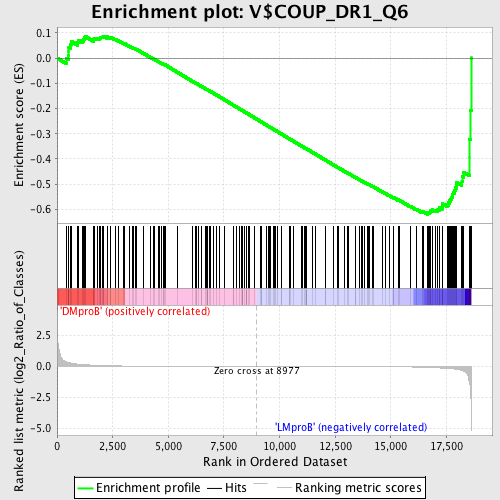
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$COUP_DR1_Q6 |
| Enrichment Score (ES) | -0.6198581 |
| Normalized Enrichment Score (NES) | -1.4564253 |
| Nominal p-value | 0.0065645515 |
| FDR q-value | 0.9257998 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKCD | 21897 | 406 | 0.366 | -0.0015 | No | ||
| 2 | RRBP1 | 8176 14411 | 522 | 0.301 | 0.0092 | No | ||
| 3 | ESRRA | 23802 | 527 | 0.299 | 0.0258 | No | ||
| 4 | ZYX | 10443 | 530 | 0.297 | 0.0423 | No | ||
| 5 | LAMP2 | 9267 2653 | 590 | 0.274 | 0.0545 | No | ||
| 6 | TCEA2 | 14699 | 644 | 0.257 | 0.0661 | No | ||
| 7 | SMAD3 | 19084 | 925 | 0.187 | 0.0614 | No | ||
| 8 | PKP3 | 18016 | 948 | 0.183 | 0.0705 | No | ||
| 9 | FGD2 | 23309 | 1132 | 0.155 | 0.0693 | No | ||
| 10 | ARF6 | 21252 | 1189 | 0.147 | 0.0745 | No | ||
| 11 | PGF | 21020 | 1215 | 0.143 | 0.0812 | No | ||
| 12 | AGPS | 14979 | 1272 | 0.136 | 0.0858 | No | ||
| 13 | USP52 | 3397 19841 | 1642 | 0.098 | 0.0713 | No | ||
| 14 | TRAF4 | 10217 5796 1400 | 1656 | 0.097 | 0.0760 | No | ||
| 15 | DMD | 24295 2647 | 1698 | 0.093 | 0.0790 | No | ||
| 16 | RBBP6 | 5365 | 1823 | 0.084 | 0.0770 | No | ||
| 17 | PCTK1 | 24369 | 1891 | 0.080 | 0.0779 | No | ||
| 18 | EIF4EBP2 | 4662 | 1929 | 0.077 | 0.0802 | No | ||
| 19 | BIN3 | 21969 | 1951 | 0.076 | 0.0833 | No | ||
| 20 | ZSWIM3 | 14733 | 2024 | 0.071 | 0.0834 | No | ||
| 21 | MAP3K11 | 11163 | 2083 | 0.067 | 0.0840 | No | ||
| 22 | RBMS1 | 14580 | 2099 | 0.065 | 0.0869 | No | ||
| 23 | SLC26A6 | 19304 | 2245 | 0.058 | 0.0823 | No | ||
| 24 | NR2F1 | 7015 21401 | 2269 | 0.057 | 0.0843 | No | ||
| 25 | LRP1 | 9284 | 2391 | 0.052 | 0.0806 | No | ||
| 26 | CLDN15 | 16667 3572 | 2403 | 0.051 | 0.0829 | No | ||
| 27 | GLG1 | 5421 | 2602 | 0.043 | 0.0746 | No | ||
| 28 | HSD17B8 | 9061 | 2760 | 0.038 | 0.0682 | No | ||
| 29 | RQCD1 | 12205 | 2973 | 0.032 | 0.0585 | No | ||
| 30 | SFRS6 | 14751 | 3050 | 0.031 | 0.0561 | No | ||
| 31 | HIP2 | 3626 3485 16838 | 3260 | 0.026 | 0.0462 | No | ||
| 32 | SS18 | 6506 6505 2030 | 3378 | 0.024 | 0.0413 | No | ||
| 33 | NCDN | 15756 2487 6471 | 3455 | 0.023 | 0.0384 | No | ||
| 34 | OBSCN | 20434 | 3536 | 0.022 | 0.0353 | No | ||
| 35 | PRRX2 | 15047 | 3588 | 0.021 | 0.0337 | No | ||
| 36 | LCAT | 18762 | 3889 | 0.017 | 0.0184 | No | ||
| 37 | PRODH2 | 1854 18300 | 4207 | 0.014 | 0.0021 | No | ||
| 38 | RNF5 | 63 | 4349 | 0.013 | -0.0048 | No | ||
| 39 | RNF139 | 7992 13291 | 4368 | 0.013 | -0.0051 | No | ||
| 40 | PDZK1 | 15490 | 4536 | 0.012 | -0.0134 | No | ||
| 41 | HSPE1 | 9133 | 4607 | 0.012 | -0.0166 | No | ||
| 42 | UBE2R2 | 16240 | 4685 | 0.011 | -0.0201 | No | ||
| 43 | ZIC4 | 5992 | 4761 | 0.011 | -0.0236 | No | ||
| 44 | OVOL1 | 23983 | 4802 | 0.011 | -0.0251 | No | ||
| 45 | YBX2 | 20818 | 4848 | 0.010 | -0.0270 | No | ||
| 46 | CYP26A1 | 23871 3695 | 4866 | 0.010 | -0.0273 | No | ||
| 47 | FOXA1 | 21060 | 5421 | 0.008 | -0.0569 | No | ||
| 48 | SALL1 | 18796 12207 | 6096 | 0.006 | -0.0931 | No | ||
| 49 | KIRREL3 | 12571 | 6212 | 0.006 | -0.0990 | No | ||
| 50 | SAP18 | 5406 | 6245 | 0.006 | -0.1004 | No | ||
| 51 | PALM | 19960 3350 | 6269 | 0.005 | -0.1014 | No | ||
| 52 | ADAMTS19 | 23544 | 6366 | 0.005 | -0.1063 | No | ||
| 53 | NR2F2 | 8619 409 | 6497 | 0.005 | -0.1131 | No | ||
| 54 | PRDM13 | 15933 | 6498 | 0.005 | -0.1128 | No | ||
| 55 | FBLN1 | 4716 2208 | 6671 | 0.004 | -0.1219 | No | ||
| 56 | CXCL14 | 21445 7165 | 6736 | 0.004 | -0.1251 | No | ||
| 57 | REST | 16818 | 6743 | 0.004 | -0.1252 | No | ||
| 58 | TOMM70A | 6608 | 6756 | 0.004 | -0.1256 | No | ||
| 59 | PPP1R14D | 14471 | 6770 | 0.004 | -0.1261 | No | ||
| 60 | STMN2 | 15638 | 6864 | 0.004 | -0.1309 | No | ||
| 61 | PRDM16 | 12893 | 6904 | 0.004 | -0.1328 | No | ||
| 62 | GOLGA4 | 12022 | 7030 | 0.004 | -0.1393 | No | ||
| 63 | GABARAPL2 | 8211 13552 | 7178 | 0.003 | -0.1471 | No | ||
| 64 | CNTFR | 2515 15906 | 7282 | 0.003 | -0.1525 | No | ||
| 65 | PIPOX | 20340 | 7535 | 0.003 | -0.1660 | No | ||
| 66 | KLK6 | 9231 9626 | 7914 | 0.002 | -0.1864 | No | ||
| 67 | GDNF | 22514 2275 | 7937 | 0.002 | -0.1875 | No | ||
| 68 | UBE2D3 | 7253 | 8043 | 0.002 | -0.1931 | No | ||
| 69 | IRX6 | 7225 | 8189 | 0.001 | -0.2009 | No | ||
| 70 | TPCN1 | 16398 6383 10915 | 8277 | 0.001 | -0.2055 | No | ||
| 71 | NRAP | 23642 | 8325 | 0.001 | -0.2080 | No | ||
| 72 | TOM1L2 | 10115 | 8337 | 0.001 | -0.2086 | No | ||
| 73 | NR4A3 | 9473 16212 5183 | 8351 | 0.001 | -0.2092 | No | ||
| 74 | THPO | 22636 | 8404 | 0.001 | -0.2120 | No | ||
| 75 | TTR | 23615 | 8504 | 0.001 | -0.2173 | No | ||
| 76 | MAP2K3 | 20856 | 8519 | 0.001 | -0.2180 | No | ||
| 77 | PRDM1 | 19775 3337 | 8528 | 0.001 | -0.2184 | No | ||
| 78 | IBRDC2 | 5747 | 8612 | 0.001 | -0.2229 | No | ||
| 79 | FOXA3 | 17949 | 8664 | 0.001 | -0.2256 | No | ||
| 80 | CRABP2 | 15554 | 8886 | 0.000 | -0.2376 | No | ||
| 81 | HMGCS2 | 15483 | 9161 | -0.000 | -0.2524 | No | ||
| 82 | CLCNKA | 15690 | 9164 | -0.000 | -0.2525 | No | ||
| 83 | HOXD3 | 9115 | 9206 | -0.000 | -0.2547 | No | ||
| 84 | PAPLN | 9301 | 9416 | -0.001 | -0.2660 | No | ||
| 85 | KCNE1L | 24044 | 9501 | -0.001 | -0.2705 | No | ||
| 86 | SLC26A3 | 21298 | 9549 | -0.001 | -0.2730 | No | ||
| 87 | ARHGEF7 | 3823 | 9605 | -0.001 | -0.2759 | No | ||
| 88 | EFNB3 | 20391 | 9703 | -0.001 | -0.2811 | No | ||
| 89 | APEX2 | 8081 | 9785 | -0.001 | -0.2854 | No | ||
| 90 | WTAP | 7215 | 9805 | -0.001 | -0.2864 | No | ||
| 91 | NR2C1 | 10215 | 9908 | -0.001 | -0.2918 | No | ||
| 92 | PAX6 | 5223 | 10071 | -0.002 | -0.3005 | No | ||
| 93 | POU5F1 | 1547 23255 | 10092 | -0.002 | -0.3015 | No | ||
| 94 | DNAJA2 | 12133 18806 | 10432 | -0.002 | -0.3197 | No | ||
| 95 | NR4A1 | 9099 | 10437 | -0.002 | -0.3198 | No | ||
| 96 | TNRC4 | 15514 | 10496 | -0.003 | -0.3228 | No | ||
| 97 | F12 | 3260 21453 | 10610 | -0.003 | -0.3288 | No | ||
| 98 | CSF2 | 20454 | 10990 | -0.003 | -0.3491 | No | ||
| 99 | TREX1 | 10219 3111 | 11032 | -0.004 | -0.3511 | No | ||
| 100 | FABP1 | 17420 | 11124 | -0.004 | -0.3558 | No | ||
| 101 | PDLIM7 | 7434 | 11156 | -0.004 | -0.3573 | No | ||
| 102 | APOM | 23007 | 11196 | -0.004 | -0.3592 | No | ||
| 103 | HOXA3 | 9108 1075 | 11197 | -0.004 | -0.3590 | No | ||
| 104 | RFX1 | 18548 | 11207 | -0.004 | -0.3593 | No | ||
| 105 | CLCN2 | 22637 | 11483 | -0.004 | -0.3739 | No | ||
| 106 | GPR124 | 18643 918 | 11601 | -0.005 | -0.3800 | No | ||
| 107 | NDUFS1 | 13940 | 12045 | -0.006 | -0.4037 | No | ||
| 108 | INSR | 18950 | 12083 | -0.006 | -0.4053 | No | ||
| 109 | PAX7 | 15697 | 12415 | -0.007 | -0.4229 | No | ||
| 110 | SLC39A5 | 3364 19596 | 12606 | -0.007 | -0.4328 | No | ||
| 111 | EFEMP1 | 20937 | 12642 | -0.007 | -0.4343 | No | ||
| 112 | SLC16A9 | 19993 | 12912 | -0.008 | -0.4484 | No | ||
| 113 | PAPD4 | 8299 8300 4146 | 13048 | -0.009 | -0.4552 | No | ||
| 114 | HOXC13 | 22344 | 13107 | -0.009 | -0.4579 | No | ||
| 115 | PRKACA | 18549 3844 | 13408 | -0.010 | -0.4736 | No | ||
| 116 | RTN4 | 7569 1328 1343 1442 | 13433 | -0.010 | -0.4743 | No | ||
| 117 | ERBB2 | 8913 | 13586 | -0.011 | -0.4819 | No | ||
| 118 | TTYH1 | 2496 7176 | 13685 | -0.012 | -0.4866 | No | ||
| 119 | SHC1 | 9813 9812 5430 | 13709 | -0.012 | -0.4871 | No | ||
| 120 | ACTA1 | 18715 | 13818 | -0.013 | -0.4923 | No | ||
| 121 | PNOC | 21783 | 13929 | -0.013 | -0.4975 | No | ||
| 122 | FXYD1 | 17874 85 | 14006 | -0.014 | -0.5009 | No | ||
| 123 | TPM2 | 2367 2437 | 14025 | -0.014 | -0.5011 | No | ||
| 124 | SYTL2 | 18190 1539 3730 | 14170 | -0.015 | -0.5080 | No | ||
| 125 | ENO3 | 8905 | 14203 | -0.015 | -0.5089 | No | ||
| 126 | CLSPN | 16079 11256 6530 | 14617 | -0.018 | -0.5303 | No | ||
| 127 | SLC23A3 | 10367 | 14760 | -0.020 | -0.5369 | No | ||
| 128 | PEX16 | 2681 | 14927 | -0.022 | -0.5446 | No | ||
| 129 | SLC11A2 | 22130 | 15115 | -0.025 | -0.5534 | No | ||
| 130 | PTHR1 | 5318 18985 | 15137 | -0.025 | -0.5531 | No | ||
| 131 | DPF3 | 7659 12853 | 15325 | -0.028 | -0.5617 | No | ||
| 132 | AGPAT1 | 23275 | 15378 | -0.029 | -0.5629 | No | ||
| 133 | ARFGEF2 | 14728 | 15861 | -0.042 | -0.5867 | No | ||
| 134 | ATP5O | 22539 | 16134 | -0.052 | -0.5985 | No | ||
| 135 | NR2F6 | 4651 8876 18848 8912 | 16411 | -0.065 | -0.6098 | No | ||
| 136 | PPARGC1A | 16533 | 16471 | -0.068 | -0.6092 | No | ||
| 137 | MYH10 | 8077 | 16669 | -0.079 | -0.6154 | Yes | ||
| 138 | SMYD5 | 17386 | 16682 | -0.081 | -0.6115 | Yes | ||
| 139 | GBL | 23091 1612 | 16757 | -0.086 | -0.6107 | Yes | ||
| 140 | STOML2 | 15902 | 16775 | -0.088 | -0.6067 | Yes | ||
| 141 | PAK4 | 17909 | 16798 | -0.090 | -0.6028 | Yes | ||
| 142 | MAGMAS | 1677 22678 | 16871 | -0.094 | -0.6015 | Yes | ||
| 143 | USP37 | 13923 | 16997 | -0.104 | -0.6024 | Yes | ||
| 144 | WBSCR16 | 16355 | 17104 | -0.112 | -0.6018 | Yes | ||
| 145 | AHCYL1 | 6037 | 17170 | -0.119 | -0.5987 | Yes | ||
| 146 | ETFDH | 15313 | 17185 | -0.121 | -0.5927 | Yes | ||
| 147 | MRPS28 | 15377 | 17304 | -0.134 | -0.5915 | Yes | ||
| 148 | DCTN1 | 1100 17392 | 17316 | -0.135 | -0.5845 | Yes | ||
| 149 | PRKAG1 | 22135 | 17334 | -0.137 | -0.5777 | Yes | ||
| 150 | ALPI | 4892 | 17531 | -0.158 | -0.5795 | Yes | ||
| 151 | PRKCSH | 19535 | 17597 | -0.167 | -0.5736 | Yes | ||
| 152 | POLR2H | 10888 | 17623 | -0.171 | -0.5653 | Yes | ||
| 153 | PSMF1 | 6009 | 17687 | -0.178 | -0.5587 | Yes | ||
| 154 | PFN1 | 9555 | 17745 | -0.186 | -0.5514 | Yes | ||
| 155 | HDAC11 | 17356 | 17761 | -0.189 | -0.5416 | Yes | ||
| 156 | HSPD1 | 4078 9129 | 17794 | -0.194 | -0.5324 | Yes | ||
| 157 | AP1B1 | 8599 | 17866 | -0.206 | -0.5247 | Yes | ||
| 158 | MRPL11 | 23774 | 17898 | -0.214 | -0.5144 | Yes | ||
| 159 | COMMD3 | 73 | 17938 | -0.223 | -0.5040 | Yes | ||
| 160 | EEF1B2 | 4131 12063 | 17949 | -0.226 | -0.4918 | Yes | ||
| 161 | ZBTB12 | 23268 | 18198 | -0.321 | -0.4873 | Yes | ||
| 162 | PCBP4 | 12235 | 18219 | -0.328 | -0.4699 | Yes | ||
| 163 | HIBADH | 17144 | 18270 | -0.367 | -0.4520 | Yes | ||
| 164 | LMAN2L | 5670 10070 5669 | 18532 | -1.291 | -0.3937 | Yes | ||
| 165 | SCOTIN | 3136 19299 3102 | 18533 | -1.296 | -0.3210 | Yes | ||
| 166 | TP53 | 20822 | 18592 | -2.086 | -0.2070 | Yes | ||
| 167 | DCTN2 | 7635 12812 | 18611 | -3.709 | 0.0003 | Yes |

