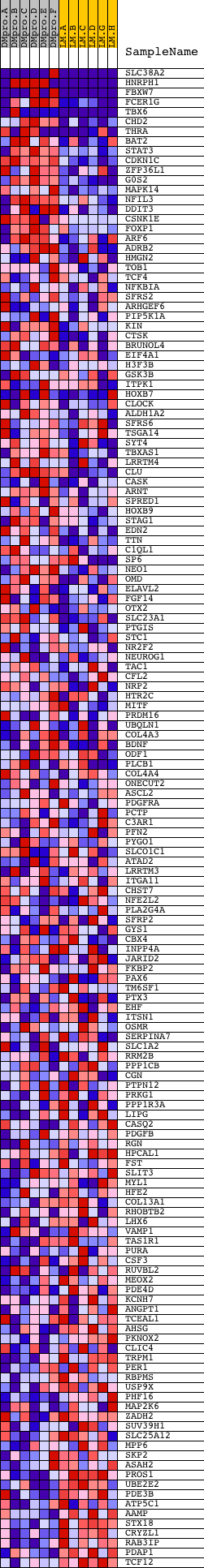
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | V$CEBP_Q2 |
| Enrichment Score (ES) | 0.6950483 |
| Normalized Enrichment Score (NES) | 1.6056192 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.7334898 |
| FWER p-Value | 0.522 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC38A2 | 22149 2219 | 4 | 4.197 | 0.2047 | Yes | ||
| 2 | HNRPH1 | 12222 1486 1220 7202 7201 | 11 | 2.956 | 0.3487 | Yes | ||
| 3 | FBXW7 | 1805 11928 | 33 | 2.037 | 0.4470 | Yes | ||
| 4 | FCER1G | 13759 | 38 | 1.883 | 0.5387 | Yes | ||
| 5 | TBX6 | 1993 18075 | 168 | 0.775 | 0.5696 | Yes | ||
| 6 | CHD2 | 10847 | 290 | 0.478 | 0.5864 | Yes | ||
| 7 | THRA | 1447 10171 1406 | 308 | 0.456 | 0.6077 | Yes | ||
| 8 | BAT2 | 23006 | 426 | 0.354 | 0.6187 | Yes | ||
| 9 | STAT3 | 5525 9906 | 448 | 0.338 | 0.6341 | Yes | ||
| 10 | CDKN1C | 17546 | 488 | 0.319 | 0.6476 | Yes | ||
| 11 | ZFP36L1 | 4453 4454 | 505 | 0.312 | 0.6619 | Yes | ||
| 12 | G0S2 | 13711 | 528 | 0.298 | 0.6753 | Yes | ||
| 13 | MAPK14 | 23313 | 659 | 0.251 | 0.6805 | Yes | ||
| 14 | NFIL3 | 21463 | 1027 | 0.171 | 0.6690 | Yes | ||
| 15 | DDIT3 | 19857 | 1073 | 0.163 | 0.6745 | Yes | ||
| 16 | CSNK1E | 6570 2211 11332 | 1179 | 0.148 | 0.6760 | Yes | ||
| 17 | FOXP1 | 4242 | 1181 | 0.148 | 0.6832 | Yes | ||
| 18 | ARF6 | 21252 | 1189 | 0.147 | 0.6900 | Yes | ||
| 19 | ADRB2 | 23422 | 1225 | 0.143 | 0.6950 | Yes | ||
| 20 | HMGN2 | 9095 | 1595 | 0.102 | 0.6800 | No | ||
| 21 | TOB1 | 20703 | 1605 | 0.101 | 0.6845 | No | ||
| 22 | TCF4 | 5642 | 1612 | 0.100 | 0.6891 | No | ||
| 23 | NFKBIA | 21065 | 1727 | 0.091 | 0.6873 | No | ||
| 24 | SFRS2 | 9807 20136 | 1747 | 0.089 | 0.6907 | No | ||
| 25 | ARHGEF6 | 13297 13104 | 1844 | 0.083 | 0.6895 | No | ||
| 26 | PIP5K1A | 23708 | 1916 | 0.078 | 0.6895 | No | ||
| 27 | KIN | 15120 | 2008 | 0.072 | 0.6881 | No | ||
| 28 | CTSK | 15506 | 2206 | 0.060 | 0.6803 | No | ||
| 29 | BRUNOL4 | 2010 4229 | 2337 | 0.054 | 0.6759 | No | ||
| 30 | EIF4A1 | 8889 23719 | 2529 | 0.046 | 0.6678 | No | ||
| 31 | H3F3B | 4839 | 2645 | 0.042 | 0.6636 | No | ||
| 32 | GSK3B | 22761 | 2764 | 0.038 | 0.6591 | No | ||
| 33 | ITPK1 | 10150 | 2845 | 0.035 | 0.6565 | No | ||
| 34 | HOXB7 | 666 9110 | 2905 | 0.034 | 0.6549 | No | ||
| 35 | CLOCK | 8749 16507 4530 | 2982 | 0.032 | 0.6524 | No | ||
| 36 | ALDH1A2 | 19386 | 2986 | 0.032 | 0.6538 | No | ||
| 37 | SFRS6 | 14751 | 3050 | 0.031 | 0.6518 | No | ||
| 38 | TSGA14 | 17194 | 3057 | 0.030 | 0.6530 | No | ||
| 39 | SYT4 | 5566 | 3186 | 0.028 | 0.6474 | No | ||
| 40 | TBXAS1 | 17480 1105 | 3229 | 0.027 | 0.6465 | No | ||
| 41 | LRRTM4 | 17405 | 3316 | 0.025 | 0.6430 | No | ||
| 42 | CLU | 21979 | 3520 | 0.022 | 0.6331 | No | ||
| 43 | CASK | 24184 2618 2604 | 3528 | 0.022 | 0.6338 | No | ||
| 44 | ARNT | 4413 1857 | 3772 | 0.019 | 0.6215 | No | ||
| 45 | SPRED1 | 4331 | 3849 | 0.018 | 0.6183 | No | ||
| 46 | HOXB9 | 20689 | 3875 | 0.017 | 0.6178 | No | ||
| 47 | STAG1 | 5520 9905 2987 | 4134 | 0.015 | 0.6045 | No | ||
| 48 | EDN2 | 16102 | 4162 | 0.015 | 0.6038 | No | ||
| 49 | TTN | 14552 2743 14553 | 4267 | 0.014 | 0.5988 | No | ||
| 50 | C1QL1 | 20201 | 4285 | 0.014 | 0.5986 | No | ||
| 51 | SP6 | 8178 13678 13679 | 4912 | 0.010 | 0.5652 | No | ||
| 52 | NEO1 | 5161 | 4936 | 0.010 | 0.5644 | No | ||
| 53 | OMD | 21643 | 5160 | 0.009 | 0.5528 | No | ||
| 54 | ELAVL2 | 15839 | 5650 | 0.007 | 0.5267 | No | ||
| 55 | FGF14 | 21716 3005 3463 | 5695 | 0.007 | 0.5246 | No | ||
| 56 | OTX2 | 9519 | 5727 | 0.007 | 0.5233 | No | ||
| 57 | SLC23A1 | 23463 | 5798 | 0.007 | 0.5198 | No | ||
| 58 | PTGIS | 9653 | 6174 | 0.006 | 0.4998 | No | ||
| 59 | STC1 | 21974 | 6380 | 0.005 | 0.4890 | No | ||
| 60 | NR2F2 | 8619 409 | 6497 | 0.005 | 0.4829 | No | ||
| 61 | NEUROG1 | 21446 | 6580 | 0.005 | 0.4787 | No | ||
| 62 | TAC1 | 17526 19853 | 6612 | 0.005 | 0.4772 | No | ||
| 63 | CFL2 | 2155 21069 | 6622 | 0.005 | 0.4770 | No | ||
| 64 | NRP2 | 5194 9486 5195 | 6758 | 0.004 | 0.4699 | No | ||
| 65 | HTR2C | 24230 | 6777 | 0.004 | 0.4691 | No | ||
| 66 | MITF | 17349 | 6811 | 0.004 | 0.4675 | No | ||
| 67 | PRDM16 | 12893 | 6904 | 0.004 | 0.4627 | No | ||
| 68 | UBQLN1 | 3237 3209 3248 3291 | 6916 | 0.004 | 0.4623 | No | ||
| 69 | COL4A3 | 8766 | 7015 | 0.004 | 0.4572 | No | ||
| 70 | BDNF | 14926 2797 | 7067 | 0.004 | 0.4546 | No | ||
| 71 | ODF1 | 22488 | 7247 | 0.003 | 0.4451 | No | ||
| 72 | PLCB1 | 14832 2821 | 7437 | 0.003 | 0.4350 | No | ||
| 73 | COL4A4 | 8767 | 7442 | 0.003 | 0.4349 | No | ||
| 74 | ONECUT2 | 10346 | 7447 | 0.003 | 0.4348 | No | ||
| 75 | ASCL2 | 5079 | 7533 | 0.003 | 0.4303 | No | ||
| 76 | PDGFRA | 16824 | 8042 | 0.002 | 0.4029 | No | ||
| 77 | PCTP | 9540 | 8162 | 0.001 | 0.3965 | No | ||
| 78 | C3AR1 | 8668 | 8275 | 0.001 | 0.3905 | No | ||
| 79 | PFN2 | 15337 1818 | 8295 | 0.001 | 0.3895 | No | ||
| 80 | PYGO1 | 19383 | 8415 | 0.001 | 0.3831 | No | ||
| 81 | SLCO1C1 | 17251 | 8737 | 0.000 | 0.3658 | No | ||
| 82 | ATAD2 | 2268 2256 | 8840 | 0.000 | 0.3603 | No | ||
| 83 | LRRTM3 | 19744 | 8922 | 0.000 | 0.3559 | No | ||
| 84 | ITGA11 | 6677 11449 | 8944 | 0.000 | 0.3547 | No | ||
| 85 | CHST7 | 24372 | 9048 | -0.000 | 0.3492 | No | ||
| 86 | NFE2L2 | 2898 14557 | 9171 | -0.000 | 0.3426 | No | ||
| 87 | PLA2G4A | 13809 | 9258 | -0.001 | 0.3379 | No | ||
| 88 | SFRP2 | 15564 | 9432 | -0.001 | 0.3286 | No | ||
| 89 | GYS1 | 4818 | 9462 | -0.001 | 0.3271 | No | ||
| 90 | CBX4 | 20128 | 9551 | -0.001 | 0.3224 | No | ||
| 91 | INPP4A | 3985 3935 11237 4085 | 9636 | -0.001 | 0.3179 | No | ||
| 92 | JARID2 | 9200 | 9651 | -0.001 | 0.3172 | No | ||
| 93 | FKBP2 | 8972 | 9977 | -0.002 | 0.2996 | No | ||
| 94 | PAX6 | 5223 | 10071 | -0.002 | 0.2947 | No | ||
| 95 | TM6SF1 | 8416 | 10631 | -0.003 | 0.2645 | No | ||
| 96 | PTX3 | 9668 15575 | 10647 | -0.003 | 0.2639 | No | ||
| 97 | EHF | 4657 | 10662 | -0.003 | 0.2633 | No | ||
| 98 | ITSN1 | 9196 | 10703 | -0.003 | 0.2612 | No | ||
| 99 | OSMR | 22333 | 10741 | -0.003 | 0.2594 | No | ||
| 100 | SERPINA7 | 6856 | 10778 | -0.003 | 0.2576 | No | ||
| 101 | SLC1A2 | 5453 | 11022 | -0.004 | 0.2446 | No | ||
| 102 | RRM2B | 6902 6903 11801 | 11023 | -0.004 | 0.2448 | No | ||
| 103 | PPP1CB | 5282 3529 3504 | 11144 | -0.004 | 0.2385 | No | ||
| 104 | CGN | 12896 12897 7701 | 11194 | -0.004 | 0.2360 | No | ||
| 105 | PTPN12 | 16597 3502 3586 3665 | 11440 | -0.004 | 0.2229 | No | ||
| 106 | PRKG1 | 5289 | 11612 | -0.005 | 0.2139 | No | ||
| 107 | PPP1R3A | 17214 | 11792 | -0.005 | 0.2045 | No | ||
| 108 | LIPG | 23407 | 11878 | -0.005 | 0.2001 | No | ||
| 109 | CASQ2 | 15477 | 11880 | -0.005 | 0.2003 | No | ||
| 110 | PDGFB | 22199 2311 | 11888 | -0.005 | 0.2002 | No | ||
| 111 | RGN | 24371 | 12375 | -0.007 | 0.1742 | No | ||
| 112 | HPCAL1 | 21307 2123 | 12553 | -0.007 | 0.1650 | No | ||
| 113 | FST | 8985 | 12685 | -0.008 | 0.1583 | No | ||
| 114 | SLIT3 | 20925 | 12761 | -0.008 | 0.1546 | No | ||
| 115 | MYL1 | 4015 | 13087 | -0.009 | 0.1374 | No | ||
| 116 | HFE2 | 1777 15495 | 13218 | -0.009 | 0.1308 | No | ||
| 117 | COL13A1 | 8763 | 13511 | -0.011 | 0.1155 | No | ||
| 118 | RHOBTB2 | 3069 6373 10902 | 13567 | -0.011 | 0.1131 | No | ||
| 119 | LHX6 | 9275 2914 | 13604 | -0.011 | 0.1117 | No | ||
| 120 | VAMP1 | 5846 1043 | 13643 | -0.011 | 0.1102 | No | ||
| 121 | TAS1R1 | 15657 | 13654 | -0.011 | 0.1102 | No | ||
| 122 | PURA | 9670 | 13829 | -0.013 | 0.1014 | No | ||
| 123 | CSF3 | 1394 20671 | 14163 | -0.015 | 0.0841 | No | ||
| 124 | RUVBL2 | 9766 | 14696 | -0.019 | 0.0562 | No | ||
| 125 | MEOX2 | 21285 | 14800 | -0.020 | 0.0516 | No | ||
| 126 | PDE4D | 10722 6235 | 14994 | -0.023 | 0.0423 | No | ||
| 127 | KCNH7 | 2891 | 15051 | -0.024 | 0.0404 | No | ||
| 128 | ANGPT1 | 8588 2260 22303 | 15086 | -0.024 | 0.0397 | No | ||
| 129 | TCEAL1 | 24245 | 15287 | -0.027 | 0.0302 | No | ||
| 130 | AHSG | 22812 | 15292 | -0.027 | 0.0313 | No | ||
| 131 | PKNOX2 | 3090 5511 9889 | 15398 | -0.030 | 0.0271 | No | ||
| 132 | CLIC4 | 11386 11342 6620 11387 | 15570 | -0.034 | 0.0195 | No | ||
| 133 | TRPM1 | 10846 474 9395 494 | 15776 | -0.039 | 0.0103 | No | ||
| 134 | PER1 | 20827 | 15815 | -0.041 | 0.0102 | No | ||
| 135 | RBPMS | 5369 | 16056 | -0.049 | -0.0004 | No | ||
| 136 | USP9X | 10264 10265 5834 | 16170 | -0.054 | -0.0039 | No | ||
| 137 | PHF16 | 11795 6901 | 16337 | -0.062 | -0.0099 | No | ||
| 138 | MAP2K6 | 20614 1414 | 16507 | -0.070 | -0.0156 | No | ||
| 139 | ZADH2 | 23501 | 16608 | -0.075 | -0.0174 | No | ||
| 140 | SUV39H1 | 24194 | 16616 | -0.076 | -0.0140 | No | ||
| 141 | SLC25A12 | 14562 | 16829 | -0.092 | -0.0210 | No | ||
| 142 | MPP6 | 17448 | 16877 | -0.095 | -0.0189 | No | ||
| 143 | SKP2 | 6576 11337 2236 | 16901 | -0.096 | -0.0155 | No | ||
| 144 | ASAH2 | 23699 | 17089 | -0.111 | -0.0202 | No | ||
| 145 | PROS1 | 22725 | 17217 | -0.124 | -0.0210 | No | ||
| 146 | UBE2E2 | 21914 | 17277 | -0.131 | -0.0178 | No | ||
| 147 | PDE3B | 5236 | 17283 | -0.132 | -0.0117 | No | ||
| 148 | ATP5C1 | 8635 | 17381 | -0.143 | -0.0099 | No | ||
| 149 | AAMP | 10406 | 17477 | -0.151 | -0.0077 | No | ||
| 150 | STX18 | 16857 | 17954 | -0.227 | -0.0224 | No | ||
| 151 | CRYZL1 | 1644 12385 | 17994 | -0.238 | -0.0129 | No | ||
| 152 | RAB3IP | 19623 | 18027 | -0.248 | -0.0026 | No | ||
| 153 | PDAP1 | 16294 | 18121 | -0.280 | 0.0061 | No | ||
| 154 | TCF12 | 10044 3125 | 18333 | -0.423 | 0.0153 | No |