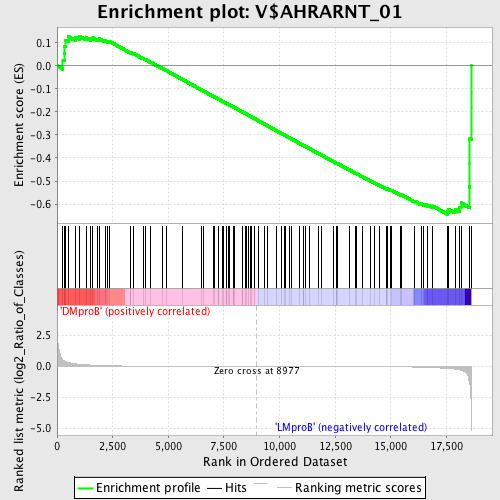
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$AHRARNT_01 |
| Enrichment Score (ES) | -0.64529026 |
| Normalized Enrichment Score (NES) | -1.4134624 |
| Nominal p-value | 0.032388665 |
| FDR q-value | 0.712831 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SRRM2 | 7999 13299 | 263 | 0.516 | 0.0241 | No | ||
| 2 | BCL11A | 4691 | 318 | 0.446 | 0.0542 | No | ||
| 3 | WSB1 | 20330 | 331 | 0.426 | 0.0852 | No | ||
| 4 | HK2 | 17106 | 375 | 0.383 | 0.1113 | No | ||
| 5 | SGK | 9809 3419 | 515 | 0.305 | 0.1264 | No | ||
| 6 | PPRC1 | 23808 | 824 | 0.207 | 0.1251 | No | ||
| 7 | XPO6 | 17644 | 994 | 0.175 | 0.1290 | No | ||
| 8 | SOX4 | 5479 | 1304 | 0.132 | 0.1221 | No | ||
| 9 | ADAM9 | 18910 | 1505 | 0.110 | 0.1195 | No | ||
| 10 | ACSL4 | 24043 11936 24042 2614 6940 | 1611 | 0.101 | 0.1212 | No | ||
| 11 | DTX1 | 16396 | 1828 | 0.084 | 0.1158 | No | ||
| 12 | RBBP7 | 10887 6366 10886 | 1903 | 0.079 | 0.1177 | No | ||
| 13 | HES1 | 22798 | 2155 | 0.063 | 0.1088 | No | ||
| 14 | HOXA5 | 17149 | 2274 | 0.057 | 0.1067 | No | ||
| 15 | HRBL | 3452 16326 3614 | 2365 | 0.053 | 0.1058 | No | ||
| 16 | SOAT1 | 13797 | 3301 | 0.026 | 0.0572 | No | ||
| 17 | REPIN1 | 12220 | 3307 | 0.025 | 0.0588 | No | ||
| 18 | IL7 | 4921 | 3448 | 0.023 | 0.0529 | No | ||
| 19 | HOXB9 | 20689 | 3875 | 0.017 | 0.0312 | No | ||
| 20 | SESN2 | 15733 | 3951 | 0.017 | 0.0284 | No | ||
| 21 | LHX1 | 4995 | 4199 | 0.014 | 0.0162 | No | ||
| 22 | PITPNC1 | 7759 | 4734 | 0.011 | -0.0118 | No | ||
| 23 | JUB | 21834 3339 | 4903 | 0.010 | -0.0202 | No | ||
| 24 | ELAVL2 | 15839 | 5650 | 0.007 | -0.0599 | No | ||
| 25 | GHR | 22336 | 6500 | 0.005 | -0.1054 | No | ||
| 26 | KAZALD1 | 4206 | 6599 | 0.005 | -0.1103 | No | ||
| 27 | GABRA1 | 20492 | 7028 | 0.004 | -0.1332 | No | ||
| 28 | BDNF | 14926 2797 | 7067 | 0.004 | -0.1349 | No | ||
| 29 | WNT4 | 16025 | 7246 | 0.003 | -0.1443 | No | ||
| 30 | GRM7 | 17338 | 7272 | 0.003 | -0.1454 | No | ||
| 31 | ONECUT2 | 10346 | 7447 | 0.003 | -0.1546 | No | ||
| 32 | PTF1A | 15110 | 7465 | 0.003 | -0.1553 | No | ||
| 33 | ZBTB8 | 15747 | 7597 | 0.003 | -0.1622 | No | ||
| 34 | NR3C1 | 9043 | 7598 | 0.003 | -0.1620 | No | ||
| 35 | CNNM1 | 23846 | 7718 | 0.002 | -0.1683 | No | ||
| 36 | CNTNAP1 | 11987 | 7767 | 0.002 | -0.1707 | No | ||
| 37 | PAX2 | 9530 24416 | 7939 | 0.002 | -0.1798 | No | ||
| 38 | GTF2A1 | 2064 8187 8188 13512 | 7944 | 0.002 | -0.1799 | No | ||
| 39 | JAG1 | 14415 | 7963 | 0.002 | -0.1807 | No | ||
| 40 | VDP | 16794 | 8310 | 0.001 | -0.1993 | No | ||
| 41 | FOXD3 | 9088 | 8326 | 0.001 | -0.2000 | No | ||
| 42 | MOV10 | 9403 9402 5114 | 8481 | 0.001 | -0.2083 | No | ||
| 43 | NEUROG2 | 15426 | 8524 | 0.001 | -0.2105 | No | ||
| 44 | GSH1 | 16622 | 8580 | 0.001 | -0.2134 | No | ||
| 45 | NAGLU | 11340 | 8685 | 0.001 | -0.2190 | No | ||
| 46 | SEZ6 | 20764 | 8728 | 0.000 | -0.2212 | No | ||
| 47 | MXI1 | 23813 5139 | 8878 | 0.000 | -0.2293 | No | ||
| 48 | GNAO1 | 4785 3829 | 9038 | -0.000 | -0.2378 | No | ||
| 49 | CRH | 8784 | 9300 | -0.001 | -0.2519 | No | ||
| 50 | KLF15 | 17359 | 9446 | -0.001 | -0.2597 | No | ||
| 51 | CTGF | 20068 | 9870 | -0.001 | -0.2824 | No | ||
| 52 | PAX6 | 5223 | 10071 | -0.002 | -0.2931 | No | ||
| 53 | SPOCK2 | 13585 | 10212 | -0.002 | -0.3005 | No | ||
| 54 | ATOH1 | 4419 | 10279 | -0.002 | -0.3039 | No | ||
| 55 | NR4A1 | 9099 | 10437 | -0.002 | -0.3122 | No | ||
| 56 | PPM2C | 491 | 10534 | -0.003 | -0.3172 | No | ||
| 57 | STC2 | 5526 | 10875 | -0.003 | -0.3353 | No | ||
| 58 | MOSPD2 | 24009 | 11083 | -0.004 | -0.3462 | No | ||
| 59 | NLGN3 | 10878 | 11093 | -0.004 | -0.3464 | No | ||
| 60 | BHLHB3 | 16935 | 11165 | -0.004 | -0.3499 | No | ||
| 61 | INSM1 | 14815 | 11359 | -0.004 | -0.3600 | No | ||
| 62 | PHPT1 | 14656 | 11741 | -0.005 | -0.3802 | No | ||
| 63 | PDGFB | 22199 2311 | 11888 | -0.005 | -0.3877 | No | ||
| 64 | RGS6 | 21219 | 12418 | -0.007 | -0.4158 | No | ||
| 65 | HPCAL1 | 21307 2123 | 12553 | -0.007 | -0.4225 | No | ||
| 66 | NEUROD2 | 20262 | 12569 | -0.007 | -0.4228 | No | ||
| 67 | RARG | 22113 | 12596 | -0.007 | -0.4236 | No | ||
| 68 | H2AFZ | 6957 | 13127 | -0.009 | -0.4516 | No | ||
| 69 | DLL4 | 7040 | 13405 | -0.010 | -0.4658 | No | ||
| 70 | TRIM23 | 8165 3230 | 13438 | -0.010 | -0.4667 | No | ||
| 71 | SHC1 | 9813 9812 5430 | 13709 | -0.012 | -0.4804 | No | ||
| 72 | RKHD3 | 18199 | 14067 | -0.014 | -0.4987 | No | ||
| 73 | MBNL1 | 1921 15582 | 14268 | -0.015 | -0.5083 | No | ||
| 74 | CAMK2D | 4232 | 14488 | -0.017 | -0.5189 | No | ||
| 75 | CYP26B1 | 17093 | 14783 | -0.020 | -0.5332 | No | ||
| 76 | PHF1 | 23324 1551 | 14820 | -0.021 | -0.5337 | No | ||
| 77 | ELAVL4 | 15805 4889 9137 | 14859 | -0.021 | -0.5342 | No | ||
| 78 | DMC1 | 22207 | 14980 | -0.023 | -0.5390 | No | ||
| 79 | SYT12 | 23962 | 15025 | -0.023 | -0.5396 | No | ||
| 80 | RBM6 | 9707 3095 3063 2995 | 15430 | -0.030 | -0.5592 | No | ||
| 81 | ELK3 | 4663 94 3388 | 15500 | -0.032 | -0.5605 | No | ||
| 82 | EGR3 | 4656 | 16068 | -0.050 | -0.5874 | No | ||
| 83 | TGFB1 | 18332 | 16358 | -0.063 | -0.5984 | No | ||
| 84 | STAM | 2912 15117 | 16487 | -0.068 | -0.6002 | No | ||
| 85 | CDK6 | 16600 | 16668 | -0.079 | -0.6041 | No | ||
| 86 | TAF11 | 12733 | 16880 | -0.095 | -0.6084 | No | ||
| 87 | MAEA | 16877 | 17564 | -0.163 | -0.6332 | Yes | ||
| 88 | RUNX1 | 4481 | 17595 | -0.167 | -0.6224 | Yes | ||
| 89 | FBXL20 | 13010 | 17887 | -0.212 | -0.6224 | Yes | ||
| 90 | UGCGL1 | 6721 | 18095 | -0.272 | -0.6134 | Yes | ||
| 91 | MAGED2 | 24034 | 18156 | -0.293 | -0.5949 | Yes | ||
| 92 | USP1 | 10497 10498 6051 | 18523 | -1.208 | -0.5250 | Yes | ||
| 93 | EIF4A2 | 4660 1679 1645 | 18549 | -1.405 | -0.4221 | Yes | ||
| 94 | TCF7L2 | 10048 5646 | 18551 | -1.410 | -0.3175 | Yes | ||
| 95 | PABPC1 | 5219 9522 9523 23572 | 18613 | -4.324 | 0.0002 | Yes |

