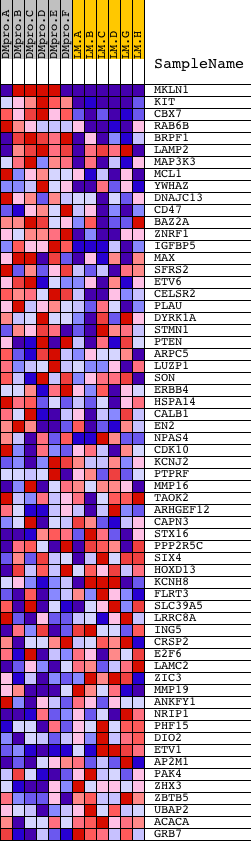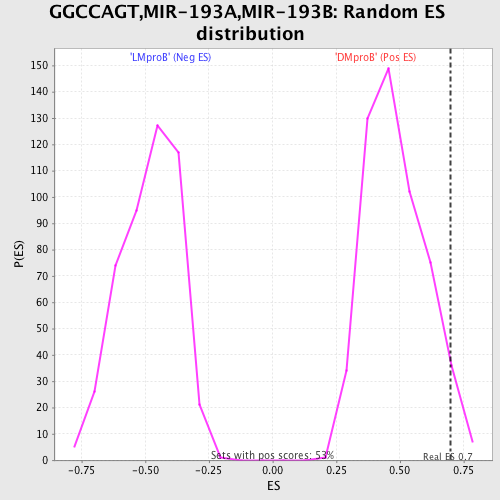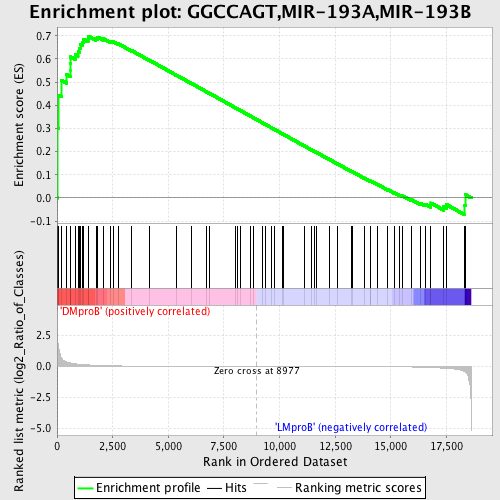
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | GGCCAGT,MIR-193A,MIR-193B |
| Enrichment Score (ES) | 0.69872504 |
| Normalized Enrichment Score (NES) | 1.4473554 |
| Nominal p-value | 0.04494382 |
| FDR q-value | 0.76876587 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MKLN1 | 6584 11339 | 12 | 2.911 | 0.3020 | Yes | ||
| 2 | KIT | 16823 | 75 | 1.382 | 0.4423 | Yes | ||
| 3 | CBX7 | 11973 | 189 | 0.691 | 0.5080 | Yes | ||
| 4 | RAB6B | 11289 | 424 | 0.355 | 0.5324 | Yes | ||
| 5 | BRPF1 | 17332 | 587 | 0.276 | 0.5523 | Yes | ||
| 6 | LAMP2 | 9267 2653 | 590 | 0.274 | 0.5807 | Yes | ||
| 7 | MAP3K3 | 20626 | 595 | 0.273 | 0.6089 | Yes | ||
| 8 | MCL1 | 15502 | 811 | 0.210 | 0.6192 | Yes | ||
| 9 | YWHAZ | 10370 | 939 | 0.186 | 0.6316 | Yes | ||
| 10 | DNAJC13 | 6177 | 990 | 0.176 | 0.6472 | Yes | ||
| 11 | CD47 | 4933 | 1038 | 0.169 | 0.6623 | Yes | ||
| 12 | BAZ2A | 4371 | 1144 | 0.154 | 0.6726 | Yes | ||
| 13 | ZNRF1 | 9306 | 1186 | 0.147 | 0.6857 | Yes | ||
| 14 | IGFBP5 | 4899 | 1391 | 0.121 | 0.6873 | Yes | ||
| 15 | MAX | 21034 | 1411 | 0.120 | 0.6987 | Yes | ||
| 16 | SFRS2 | 9807 20136 | 1747 | 0.089 | 0.6899 | No | ||
| 17 | ETV6 | 17264 | 1825 | 0.084 | 0.6945 | No | ||
| 18 | CELSR2 | 15194 12000 | 2069 | 0.067 | 0.6884 | No | ||
| 19 | PLAU | 22084 | 2417 | 0.050 | 0.6750 | No | ||
| 20 | DYRK1A | 4649 | 2521 | 0.046 | 0.6742 | No | ||
| 21 | STMN1 | 9261 | 2749 | 0.038 | 0.6659 | No | ||
| 22 | PTEN | 5305 | 3348 | 0.025 | 0.6362 | No | ||
| 23 | ARPC5 | 12580 7487 | 4151 | 0.015 | 0.5946 | No | ||
| 24 | LUZP1 | 6533 11257 2503 | 5384 | 0.008 | 0.5290 | No | ||
| 25 | SON | 5473 1657 1684 | 6024 | 0.006 | 0.4952 | No | ||
| 26 | ERBB4 | 13933 4678 | 6731 | 0.004 | 0.4576 | No | ||
| 27 | HSPA14 | 14695 | 6830 | 0.004 | 0.4528 | No | ||
| 28 | CALB1 | 4469 | 8012 | 0.002 | 0.3893 | No | ||
| 29 | EN2 | 16898 | 8096 | 0.002 | 0.3850 | No | ||
| 30 | NPAS4 | 23970 3683 | 8235 | 0.001 | 0.3777 | No | ||
| 31 | CDK10 | 3818 6157 10635 3894 | 8681 | 0.001 | 0.3537 | No | ||
| 32 | KCNJ2 | 20612 | 8819 | 0.000 | 0.3464 | No | ||
| 33 | PTPRF | 5331 2545 | 9244 | -0.000 | 0.3236 | No | ||
| 34 | MMP16 | 5109 | 9363 | -0.001 | 0.3173 | No | ||
| 35 | TAOK2 | 10610 3794 | 9634 | -0.001 | 0.3028 | No | ||
| 36 | ARHGEF12 | 19156 | 9754 | -0.001 | 0.2966 | No | ||
| 37 | CAPN3 | 8686 2868 | 10141 | -0.002 | 0.2760 | No | ||
| 38 | STX16 | 10464 | 10177 | -0.002 | 0.2743 | No | ||
| 39 | PPP2R5C | 6522 2147 | 11128 | -0.004 | 0.2235 | No | ||
| 40 | SIX4 | 5445 | 11445 | -0.004 | 0.2069 | No | ||
| 41 | HOXD13 | 9114 | 11577 | -0.005 | 0.2003 | No | ||
| 42 | KCNH8 | 23197 | 11637 | -0.005 | 0.1976 | No | ||
| 43 | FLRT3 | 7730 12930 2673 | 12249 | -0.006 | 0.1654 | No | ||
| 44 | SLC39A5 | 3364 19596 | 12606 | -0.007 | 0.1469 | No | ||
| 45 | LRRC8A | 15054 | 13248 | -0.009 | 0.1134 | No | ||
| 46 | ING5 | 12314 7273 4039 3964 | 13268 | -0.010 | 0.1133 | No | ||
| 47 | CRSP2 | 11223 | 13799 | -0.012 | 0.0861 | No | ||
| 48 | E2F6 | 6925 6926 11920 | 14085 | -0.014 | 0.0722 | No | ||
| 49 | LAMC2 | 4983 9266 13806 3975 | 14094 | -0.014 | 0.0732 | No | ||
| 50 | ZIC3 | 10430 | 14378 | -0.016 | 0.0597 | No | ||
| 51 | MMP19 | 7191 | 14867 | -0.021 | 0.0356 | No | ||
| 52 | ANKFY1 | 1405 4386 | 15156 | -0.025 | 0.0227 | No | ||
| 53 | NRIP1 | 11216 | 15405 | -0.030 | 0.0124 | No | ||
| 54 | PHF15 | 20467 8050 | 15517 | -0.032 | 0.0098 | No | ||
| 55 | DIO2 | 21014 2159 | 15946 | -0.045 | -0.0086 | No | ||
| 56 | ETV1 | 4688 8925 8924 | 16353 | -0.062 | -0.0240 | No | ||
| 57 | AP2M1 | 8603 | 16556 | -0.072 | -0.0274 | No | ||
| 58 | PAK4 | 17909 | 16798 | -0.090 | -0.0311 | No | ||
| 59 | ZHX3 | 6800 | 16803 | -0.090 | -0.0220 | No | ||
| 60 | ZBTB5 | 15891 | 17374 | -0.142 | -0.0380 | No | ||
| 61 | UBAP2 | 15912 | 17506 | -0.155 | -0.0289 | No | ||
| 62 | ACACA | 309 4209 | 18298 | -0.389 | -0.0311 | No | ||
| 63 | GRB7 | 20673 | 18363 | -0.464 | 0.0136 | No |