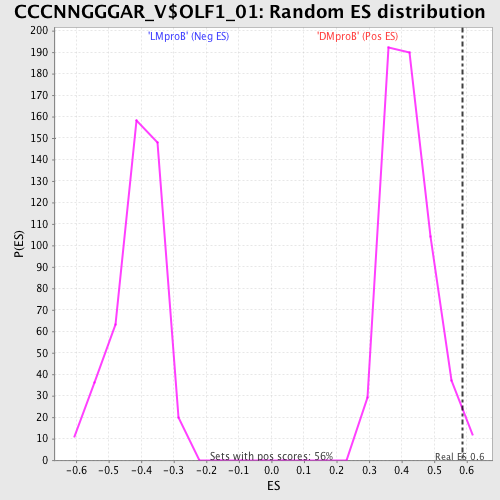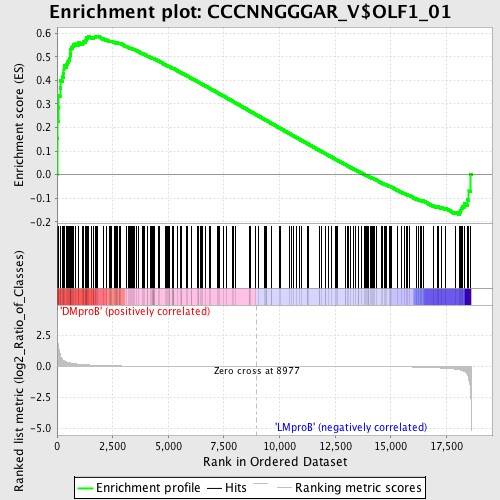
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | CCCNNGGGAR_V$OLF1_01 |
| Enrichment Score (ES) | 0.5876051 |
| Normalized Enrichment Score (NES) | 1.4052869 |
| Nominal p-value | 0.021276595 |
| FDR q-value | 0.7864681 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 3 | 4.234 | 0.1543 | Yes | ||
| 2 | TNIP1 | 12192 20450 1320 | 35 | 2.016 | 0.2262 | Yes | ||
| 3 | UBTF | 1313 10052 | 49 | 1.608 | 0.2842 | Yes | ||
| 4 | LASP1 | 4986 4985 1248 | 72 | 1.396 | 0.3339 | Yes | ||
| 5 | MAPKAP1 | 15030 5993 | 133 | 1.016 | 0.3677 | Yes | ||
| 6 | ABI3 | 7314 | 155 | 0.862 | 0.3980 | Yes | ||
| 7 | ROBO3 | 9705 | 236 | 0.567 | 0.4144 | Yes | ||
| 8 | BCL9L | 19475 | 273 | 0.499 | 0.4306 | Yes | ||
| 9 | THRA | 1447 10171 1406 | 308 | 0.456 | 0.4454 | Yes | ||
| 10 | FCHSD2 | 18172 | 311 | 0.453 | 0.4618 | Yes | ||
| 11 | BAT2 | 23006 | 426 | 0.354 | 0.4686 | Yes | ||
| 12 | GNGT2 | 20691 | 474 | 0.326 | 0.4779 | Yes | ||
| 13 | DTX3 | 19607 | 529 | 0.298 | 0.4859 | Yes | ||
| 14 | TIAM1 | 22547 | 566 | 0.283 | 0.4942 | Yes | ||
| 15 | AXL | 17922 | 579 | 0.278 | 0.5037 | Yes | ||
| 16 | TMEM24 | 19150 | 591 | 0.274 | 0.5131 | Yes | ||
| 17 | CD79B | 20185 1309 | 602 | 0.271 | 0.5225 | Yes | ||
| 18 | TRIM8 | 23824 | 603 | 0.271 | 0.5324 | Yes | ||
| 19 | HCST | 6237 | 627 | 0.262 | 0.5407 | Yes | ||
| 20 | TST | 22222 | 708 | 0.235 | 0.5449 | Yes | ||
| 21 | IL4I1 | 8971 | 730 | 0.229 | 0.5521 | Yes | ||
| 22 | UBAP1 | 7388 | 830 | 0.206 | 0.5543 | Yes | ||
| 23 | SLC25A28 | 23669 | 945 | 0.184 | 0.5548 | Yes | ||
| 24 | H1F0 | 22428 | 960 | 0.182 | 0.5607 | Yes | ||
| 25 | BAZ2A | 4371 | 1144 | 0.154 | 0.5563 | Yes | ||
| 26 | RAB10 | 2090 21119 | 1176 | 0.149 | 0.5601 | Yes | ||
| 27 | GNB2 | 9026 | 1196 | 0.146 | 0.5644 | Yes | ||
| 28 | MESDC1 | 17771 | 1278 | 0.135 | 0.5649 | Yes | ||
| 29 | HIC2 | 7185 | 1283 | 0.135 | 0.5696 | Yes | ||
| 30 | FOXJ2 | 17294 | 1319 | 0.130 | 0.5725 | Yes | ||
| 31 | POU2AF1 | 19455 | 1332 | 0.129 | 0.5765 | Yes | ||
| 32 | CHCHD7 | 16280 | 1337 | 0.127 | 0.5809 | Yes | ||
| 33 | PSCD3 | 16633 | 1379 | 0.123 | 0.5832 | Yes | ||
| 34 | RNF44 | 8364 | 1412 | 0.120 | 0.5858 | Yes | ||
| 35 | IGF2R | 23123 | 1557 | 0.105 | 0.5818 | Yes | ||
| 36 | SLC25A14 | 24344 | 1638 | 0.098 | 0.5811 | Yes | ||
| 37 | TRAF4 | 10217 5796 1400 | 1656 | 0.097 | 0.5837 | Yes | ||
| 38 | NFKBIA | 21065 | 1727 | 0.091 | 0.5832 | Yes | ||
| 39 | BMP4 | 21863 | 1731 | 0.091 | 0.5863 | Yes | ||
| 40 | UBE2N | 8216 19898 | 1769 | 0.088 | 0.5875 | Yes | ||
| 41 | ETV6 | 17264 | 1825 | 0.084 | 0.5876 | Yes | ||
| 42 | RBM10 | 6188 10677 2569 | 2065 | 0.067 | 0.5771 | No | ||
| 43 | PIAS1 | 7126 | 2211 | 0.060 | 0.5714 | No | ||
| 44 | ATF5 | 17843 | 2344 | 0.054 | 0.5662 | No | ||
| 45 | LRP1 | 9284 | 2391 | 0.052 | 0.5656 | No | ||
| 46 | CLDN15 | 16667 3572 | 2403 | 0.051 | 0.5668 | No | ||
| 47 | FLT1 | 3483 16287 | 2465 | 0.049 | 0.5653 | No | ||
| 48 | MADCAM1 | 19966 | 2591 | 0.044 | 0.5601 | No | ||
| 49 | H3F3B | 4839 | 2645 | 0.042 | 0.5587 | No | ||
| 50 | TNKS1BP1 | 14970 | 2648 | 0.042 | 0.5602 | No | ||
| 51 | ABR | 8468 20346 1476 1429 | 2651 | 0.041 | 0.5616 | No | ||
| 52 | VAT1 | 11253 | 2719 | 0.039 | 0.5593 | No | ||
| 53 | CD72 | 8718 | 2787 | 0.037 | 0.5570 | No | ||
| 54 | TNNT3 | 18004 | 2804 | 0.036 | 0.5575 | No | ||
| 55 | RAB39 | 19113 | 2850 | 0.035 | 0.5563 | No | ||
| 56 | FSTL3 | 19961 | 3103 | 0.029 | 0.5437 | No | ||
| 57 | JMJD1B | 23600 | 3216 | 0.027 | 0.5386 | No | ||
| 58 | MYOZ1 | 21907 | 3243 | 0.027 | 0.5382 | No | ||
| 59 | REPIN1 | 12220 | 3307 | 0.025 | 0.5357 | No | ||
| 60 | ANXA9 | 12964 | 3355 | 0.024 | 0.5340 | No | ||
| 61 | GRIK3 | 16083 | 3390 | 0.024 | 0.5331 | No | ||
| 62 | NCDN | 15756 2487 6471 | 3455 | 0.023 | 0.5304 | No | ||
| 63 | EPN3 | 20291 | 3465 | 0.023 | 0.5308 | No | ||
| 64 | STAG2 | 5521 | 3577 | 0.021 | 0.5255 | No | ||
| 65 | FBXL19 | 6134 | 3665 | 0.020 | 0.5215 | No | ||
| 66 | PACSIN1 | 6257 23322 10744 | 3831 | 0.018 | 0.5132 | No | ||
| 67 | BCOR | 12934 2561 | 3862 | 0.018 | 0.5122 | No | ||
| 68 | CHRDL1 | 8180 13504 | 3945 | 0.017 | 0.5084 | No | ||
| 69 | APBA1 | 11483 | 4076 | 0.015 | 0.5019 | No | ||
| 70 | HOXA11 | 17146 | 4084 | 0.015 | 0.5020 | No | ||
| 71 | ADRB1 | 4357 | 4201 | 0.014 | 0.4963 | No | ||
| 72 | PTMA | 9657 | 4223 | 0.014 | 0.4956 | No | ||
| 73 | C1QL1 | 20201 | 4285 | 0.014 | 0.4928 | No | ||
| 74 | ETV4 | 20212 | 4309 | 0.014 | 0.4921 | No | ||
| 75 | GREM1 | 14485 | 4322 | 0.013 | 0.4919 | No | ||
| 76 | DPF1 | 6616 18309 | 4327 | 0.013 | 0.4922 | No | ||
| 77 | PSME3 | 20657 | 4348 | 0.013 | 0.4916 | No | ||
| 78 | DAB1 | 4594 8834 | 4377 | 0.013 | 0.4906 | No | ||
| 79 | DPYSL5 | 92 | 4384 | 0.013 | 0.4907 | No | ||
| 80 | ELL2 | 5329 | 4565 | 0.012 | 0.4814 | No | ||
| 81 | COL23A1 | 10696 | 4606 | 0.012 | 0.4796 | No | ||
| 82 | WDR13 | 7859 13112 | 4856 | 0.010 | 0.4665 | No | ||
| 83 | NEO1 | 5161 | 4936 | 0.010 | 0.4626 | No | ||
| 84 | GJC1 | 20256 | 4942 | 0.010 | 0.4627 | No | ||
| 85 | RHOQ | 23142 | 5003 | 0.010 | 0.4598 | No | ||
| 86 | ENPP2 | 9548 | 5042 | 0.010 | 0.4580 | No | ||
| 87 | DUSP4 | 18632 3820 | 5049 | 0.010 | 0.4581 | No | ||
| 88 | LRFN3 | 17891 | 5170 | 0.009 | 0.4519 | No | ||
| 89 | LRP8 | 16149 2425 | 5196 | 0.009 | 0.4508 | No | ||
| 90 | SMPD3 | 18757 | 5219 | 0.009 | 0.4500 | No | ||
| 91 | PPP3CB | 5285 | 5226 | 0.009 | 0.4500 | No | ||
| 92 | HPCA | 9118 | 5232 | 0.009 | 0.4500 | No | ||
| 93 | SNCB | 21456 | 5426 | 0.008 | 0.4398 | No | ||
| 94 | COL11A2 | 23286 | 5536 | 0.008 | 0.4342 | No | ||
| 95 | PRRX1 | 9595 5271 | 5585 | 0.008 | 0.4319 | No | ||
| 96 | LHX2 | 15019 | 5610 | 0.007 | 0.4308 | No | ||
| 97 | TBX21 | 20276 | 5831 | 0.007 | 0.4191 | No | ||
| 98 | KCND1 | 4940 | 5866 | 0.007 | 0.4175 | No | ||
| 99 | EPHB3 | 22816 | 6030 | 0.006 | 0.4089 | No | ||
| 100 | LPP | 5570 | 6054 | 0.006 | 0.4079 | No | ||
| 101 | ATP2B3 | 24308 11557 | 6314 | 0.005 | 0.3940 | No | ||
| 102 | DUSP22 | 466 21680 | 6359 | 0.005 | 0.3918 | No | ||
| 103 | SCG3 | 19061 | 6367 | 0.005 | 0.3916 | No | ||
| 104 | NR2E1 | 19780 | 6449 | 0.005 | 0.3874 | No | ||
| 105 | IL11 | 17981 | 6489 | 0.005 | 0.3854 | No | ||
| 106 | SLC41A1 | 8269 | 6530 | 0.005 | 0.3834 | No | ||
| 107 | MYOHD1 | 20724 | 6667 | 0.004 | 0.3762 | No | ||
| 108 | CHL1 | 17347 1146 8741 4520 | 6847 | 0.004 | 0.3666 | No | ||
| 109 | ALOX12B | 20825 | 6894 | 0.004 | 0.3643 | No | ||
| 110 | MPST | 22434 | 7227 | 0.003 | 0.3464 | No | ||
| 111 | GRM7 | 17338 | 7272 | 0.003 | 0.3441 | No | ||
| 112 | PCYT1B | 24289 | 7279 | 0.003 | 0.3439 | No | ||
| 113 | VASP | 5847 | 7474 | 0.003 | 0.3335 | No | ||
| 114 | SDCCAG8 | 14034 | 7612 | 0.003 | 0.3261 | No | ||
| 115 | HR | 3622 9119 | 7862 | 0.002 | 0.3127 | No | ||
| 116 | PAX2 | 9530 24416 | 7939 | 0.002 | 0.3086 | No | ||
| 117 | LUC7L2 | 5313 5312 | 8039 | 0.002 | 0.3033 | No | ||
| 118 | POGK | 7739 | 8639 | 0.001 | 0.2708 | No | ||
| 119 | SGK2 | 14750 | 8666 | 0.001 | 0.2694 | No | ||
| 120 | MDGA1 | 13231 | 8694 | 0.000 | 0.2679 | No | ||
| 121 | LRRTM3 | 19744 | 8922 | 0.000 | 0.2556 | No | ||
| 122 | GNAO1 | 4785 3829 | 9038 | -0.000 | 0.2494 | No | ||
| 123 | WNT7B | 22172 | 9319 | -0.001 | 0.2342 | No | ||
| 124 | PTHLH | 16931 | 9355 | -0.001 | 0.2323 | No | ||
| 125 | PIGW | 20312 | 9403 | -0.001 | 0.2298 | No | ||
| 126 | SFRP2 | 15564 | 9432 | -0.001 | 0.2283 | No | ||
| 127 | MTSS1 | 5585 22285 | 9627 | -0.001 | 0.2178 | No | ||
| 128 | JARID2 | 9200 | 9651 | -0.001 | 0.2166 | No | ||
| 129 | FKBP2 | 8972 | 9977 | -0.002 | 0.1990 | No | ||
| 130 | ADCY4 | 2980 21818 | 10062 | -0.002 | 0.1945 | No | ||
| 131 | SP3 | 5483 | 10445 | -0.002 | 0.1738 | No | ||
| 132 | RHOG | 17727 | 10535 | -0.003 | 0.1691 | No | ||
| 133 | CNTN2 | 5627 | 10609 | -0.003 | 0.1652 | No | ||
| 134 | CDON | 12195 | 10738 | -0.003 | 0.1584 | No | ||
| 135 | STC2 | 5526 | 10875 | -0.003 | 0.1511 | No | ||
| 136 | TOM1L1 | 20296 1330 | 10974 | -0.003 | 0.1459 | No | ||
| 137 | SLC9A6 | 2590 6189 | 11251 | -0.004 | 0.1311 | No | ||
| 138 | RORC | 9732 | 11276 | -0.004 | 0.1299 | No | ||
| 139 | JMJD2A | 10505 6058 | 11280 | -0.004 | 0.1299 | No | ||
| 140 | PDGFC | 7048 | 11796 | -0.005 | 0.1021 | No | ||
| 141 | PLXDC2 | 15114 | 11807 | -0.005 | 0.1018 | No | ||
| 142 | TRIM29 | 19483 3061 | 11861 | -0.005 | 0.0991 | No | ||
| 143 | BANP | 18440 | 12058 | -0.006 | 0.0887 | No | ||
| 144 | THBS3 | 15542 1796 | 12082 | -0.006 | 0.0876 | No | ||
| 145 | HOXA6 | 17150 | 12184 | -0.006 | 0.0824 | No | ||
| 146 | GPM6B | 9034 | 12347 | -0.007 | 0.0738 | No | ||
| 147 | FGF11 | 20383 | 12518 | -0.007 | 0.0648 | No | ||
| 148 | NEUROD2 | 20262 | 12569 | -0.007 | 0.0624 | No | ||
| 149 | ADCYAP1 | 4348 | 12612 | -0.007 | 0.0604 | No | ||
| 150 | ATP1A1 | 15222 | 12958 | -0.008 | 0.0419 | No | ||
| 151 | CALM1 | 21184 | 12968 | -0.008 | 0.0417 | No | ||
| 152 | LIN28 | 15723 | 13032 | -0.009 | 0.0386 | No | ||
| 153 | SP4 | 20972 | 13081 | -0.009 | 0.0363 | No | ||
| 154 | EVI1 | 4689 8926 | 13186 | -0.009 | 0.0310 | No | ||
| 155 | MAGEL2 | 18221 | 13306 | -0.010 | 0.0249 | No | ||
| 156 | DLL4 | 7040 | 13405 | -0.010 | 0.0200 | No | ||
| 157 | PLAG1 | 7148 15949 12161 | 13525 | -0.011 | 0.0139 | No | ||
| 158 | ELAVL3 | 9136 | 13534 | -0.011 | 0.0139 | No | ||
| 159 | TTYH1 | 2496 7176 | 13685 | -0.012 | 0.0061 | No | ||
| 160 | MLLT6 | 20678 | 13812 | -0.013 | -0.0002 | No | ||
| 161 | EFS | 21831 | 13874 | -0.013 | -0.0031 | No | ||
| 162 | NDFIP1 | 23573 | 13919 | -0.013 | -0.0050 | No | ||
| 163 | COL18A1 | 19719 3360 | 13956 | -0.013 | -0.0065 | No | ||
| 164 | MAG | 17880 | 13977 | -0.014 | -0.0071 | No | ||
| 165 | ADAMTS4 | 10791 | 13999 | -0.014 | -0.0077 | No | ||
| 166 | GCNT2 | 9168 3166 3243 21661 3270 | 14068 | -0.014 | -0.0109 | No | ||
| 167 | CHAT | 21882 | 14131 | -0.014 | -0.0137 | No | ||
| 168 | SYTL2 | 18190 1539 3730 | 14170 | -0.015 | -0.0152 | No | ||
| 169 | DDR1 | 1625 23000 | 14206 | -0.015 | -0.0166 | No | ||
| 170 | GRIK1 | 22550 1741 | 14261 | -0.015 | -0.0190 | No | ||
| 171 | ICAM1 | 19545 | 14337 | -0.016 | -0.0225 | No | ||
| 172 | BAI2 | 2541 2379 16067 | 14573 | -0.018 | -0.0346 | No | ||
| 173 | GRIN2D | 9042 | 14626 | -0.019 | -0.0367 | No | ||
| 174 | C1QTNF1 | 20579 | 14703 | -0.019 | -0.0401 | No | ||
| 175 | SLC30A5 | 3190 21361 | 14738 | -0.020 | -0.0413 | No | ||
| 176 | NF2 | 1222 5166 | 14817 | -0.020 | -0.0448 | No | ||
| 177 | CKM | 18362 | 14921 | -0.022 | -0.0496 | No | ||
| 178 | GRK5 | 9036 23814 | 14977 | -0.023 | -0.0517 | No | ||
| 179 | ECM2 | 300 | 14991 | -0.023 | -0.0516 | No | ||
| 180 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 15048 | -0.024 | -0.0538 | No | ||
| 181 | AKAP12 | 17600 | 15298 | -0.028 | -0.0663 | No | ||
| 182 | ELK3 | 4663 94 3388 | 15500 | -0.032 | -0.0760 | No | ||
| 183 | HOXC10 | 22342 | 15612 | -0.035 | -0.0808 | No | ||
| 184 | SMARCE1 | 1218 20253 | 15701 | -0.037 | -0.0842 | No | ||
| 185 | LRRC20 | 20009 | 15759 | -0.039 | -0.0859 | No | ||
| 186 | HEBP2 | 19812 | 15821 | -0.041 | -0.0877 | No | ||
| 187 | TENC1 | 22349 9927 | 16132 | -0.052 | -0.1027 | No | ||
| 188 | UPF3B | 24171 | 16235 | -0.057 | -0.1061 | No | ||
| 189 | ARMCX3 | 12951 | 16345 | -0.062 | -0.1098 | No | ||
| 190 | NUP62 | 9497 | 16359 | -0.063 | -0.1082 | No | ||
| 191 | SSH2 | 6202 | 16461 | -0.067 | -0.1112 | No | ||
| 192 | ICA1 | 17217 | 16904 | -0.097 | -0.1317 | No | ||
| 193 | EGR1 | 23598 | 17077 | -0.110 | -0.1371 | No | ||
| 194 | TPM1 | 19070 | 17146 | -0.117 | -0.1365 | No | ||
| 195 | ZDHHC14 | 5886 | 17281 | -0.131 | -0.1390 | No | ||
| 196 | AAMP | 10406 | 17477 | -0.151 | -0.1441 | No | ||
| 197 | TRIB2 | 21102 | 17895 | -0.213 | -0.1589 | No | ||
| 198 | DLX1 | 14988 | 18086 | -0.269 | -0.1594 | No | ||
| 199 | PDAP1 | 16294 | 18121 | -0.280 | -0.1511 | No | ||
| 200 | MAGED2 | 24034 | 18156 | -0.293 | -0.1422 | No | ||
| 201 | PCBP4 | 12235 | 18219 | -0.328 | -0.1336 | No | ||
| 202 | NDUFS2 | 4089 13760 | 18299 | -0.389 | -0.1237 | No | ||
| 203 | LIMK2 | 9278 1296 | 18453 | -0.686 | -0.1070 | No | ||
| 204 | MTX1 | 15282 1918 | 18512 | -1.111 | -0.0695 | No | ||
| 205 | CA2 | 15631 1825 | 18589 | -2.059 | 0.0015 | No |