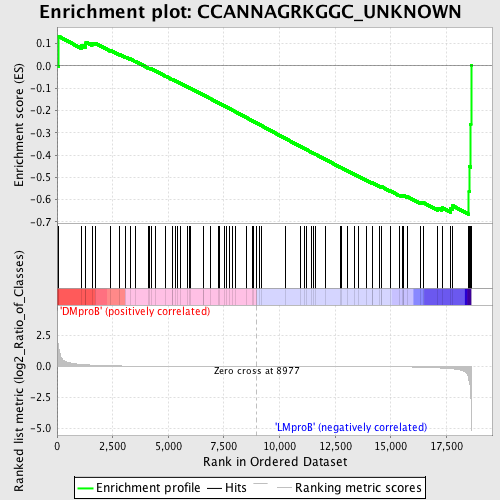
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | CCANNAGRKGGC_UNKNOWN |
| Enrichment Score (ES) | -0.66691256 |
| Normalized Enrichment Score (NES) | -1.4347929 |
| Nominal p-value | 0.03167421 |
| FDR q-value | 0.8445155 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PUM2 | 2071 8161 | 58 | 1.485 | 0.1341 | No | ||
| 2 | GRN | 20640 | 1099 | 0.159 | 0.0927 | No | ||
| 3 | PITPNM1 | 23770 | 1263 | 0.137 | 0.0966 | No | ||
| 4 | ACBD4 | 20637 | 1295 | 0.133 | 0.1073 | No | ||
| 5 | GFI1B | 14637 | 1570 | 0.104 | 0.1021 | No | ||
| 6 | C1QBP | 20364 | 1716 | 0.092 | 0.1027 | No | ||
| 7 | LRP1 | 9284 | 2391 | 0.052 | 0.0711 | No | ||
| 8 | SORCS2 | 16553 | 2802 | 0.036 | 0.0524 | No | ||
| 9 | THRAP1 | 20309 | 3080 | 0.030 | 0.0402 | No | ||
| 10 | DAPK2 | 19395 | 3281 | 0.026 | 0.0318 | No | ||
| 11 | NOLA3 | 14914 | 3516 | 0.022 | 0.0212 | No | ||
| 12 | COL11A1 | 15442 1937 4540 8762 | 4105 | 0.015 | -0.0091 | No | ||
| 13 | STAG1 | 5520 9905 2987 | 4134 | 0.015 | -0.0092 | No | ||
| 14 | PTMA | 9657 | 4223 | 0.014 | -0.0126 | No | ||
| 15 | GPR37L1 | 13825 | 4443 | 0.013 | -0.0233 | No | ||
| 16 | SMAD7 | 23513 | 4889 | 0.010 | -0.0463 | No | ||
| 17 | SLC35E4 | 20549 | 5181 | 0.009 | -0.0612 | No | ||
| 18 | SCN3A | 14573 | 5187 | 0.009 | -0.0606 | No | ||
| 19 | OTOP2 | 20600 | 5320 | 0.008 | -0.0669 | No | ||
| 20 | HOXA10 | 17145 989 | 5413 | 0.008 | -0.0712 | No | ||
| 21 | ALK | 8574 | 5538 | 0.008 | -0.0771 | No | ||
| 22 | USH1G | 20153 | 5864 | 0.007 | -0.0941 | No | ||
| 23 | SOX10 | 22211 | 5962 | 0.006 | -0.0987 | No | ||
| 24 | UPK2 | 19146 | 5995 | 0.006 | -0.0999 | No | ||
| 25 | EXT2 | 14516 | 6560 | 0.005 | -0.1298 | No | ||
| 26 | KAZALD1 | 4206 | 6599 | 0.005 | -0.1315 | No | ||
| 27 | CABP1 | 16412 | 6896 | 0.004 | -0.1471 | No | ||
| 28 | GPR156 | 10753 22763 | 7271 | 0.003 | -0.1670 | No | ||
| 29 | PIK3R1 | 3170 | 7286 | 0.003 | -0.1674 | No | ||
| 30 | GRPR | 24015 | 7515 | 0.003 | -0.1795 | No | ||
| 31 | DLX2 | 14561 353 | 7621 | 0.002 | -0.1849 | No | ||
| 32 | CNTNAP1 | 11987 | 7767 | 0.002 | -0.1925 | No | ||
| 33 | CORIN | 16517 | 7896 | 0.002 | -0.1992 | No | ||
| 34 | PCDH12 | 7005 | 8003 | 0.002 | -0.2048 | No | ||
| 35 | KCND2 | 4941 17515 | 8518 | 0.001 | -0.2324 | No | ||
| 36 | IL6 | 16895 | 8787 | 0.000 | -0.2469 | No | ||
| 37 | MB | 22232 | 8829 | 0.000 | -0.2491 | No | ||
| 38 | ANKRD2 | 23853 | 8947 | 0.000 | -0.2554 | No | ||
| 39 | ADORA1 | 13831 | 8951 | 0.000 | -0.2555 | No | ||
| 40 | ARTN | 15783 2430 2532 | 9094 | -0.000 | -0.2632 | No | ||
| 41 | STK32B | 12255 | 9188 | -0.000 | -0.2681 | No | ||
| 42 | CSMD3 | 10735 7938 | 10271 | -0.002 | -0.3263 | No | ||
| 43 | NPDC1 | 9479 | 10955 | -0.003 | -0.3628 | No | ||
| 44 | CADPS | 11298 | 11131 | -0.004 | -0.3719 | No | ||
| 45 | KPNA6 | 4972 | 11218 | -0.004 | -0.3762 | No | ||
| 46 | YY1 | 10371 | 11447 | -0.004 | -0.3881 | No | ||
| 47 | SOST | 20210 | 11507 | -0.005 | -0.3908 | No | ||
| 48 | CHRM1 | 23943 | 11616 | -0.005 | -0.3962 | No | ||
| 49 | THBS3 | 15542 1796 | 12082 | -0.006 | -0.4208 | No | ||
| 50 | POU2F3 | 9602 | 12754 | -0.008 | -0.4562 | No | ||
| 51 | SLIT3 | 20925 | 12761 | -0.008 | -0.4559 | No | ||
| 52 | ELN | 8896 | 13056 | -0.009 | -0.4709 | No | ||
| 53 | FBXO16 | 11929 | 13378 | -0.010 | -0.4873 | No | ||
| 54 | ESR2 | 21038 | 13529 | -0.011 | -0.4944 | No | ||
| 55 | RORA | 3019 5388 | 13925 | -0.013 | -0.5145 | No | ||
| 56 | RERG | 16949 | 14160 | -0.015 | -0.5257 | No | ||
| 57 | CSF3 | 1394 20671 | 14163 | -0.015 | -0.5245 | No | ||
| 58 | SLCO3A1 | 17796 4237 8430 | 14507 | -0.018 | -0.5414 | No | ||
| 59 | GAD1 | 14993 2690 | 14570 | -0.018 | -0.5430 | No | ||
| 60 | BAI2 | 2541 2379 16067 | 14573 | -0.018 | -0.5415 | No | ||
| 61 | GRK5 | 9036 23814 | 14977 | -0.023 | -0.5611 | No | ||
| 62 | CXCL12 | 9792 1182 1031 | 15370 | -0.029 | -0.5796 | No | ||
| 63 | MDK | 14523 | 15523 | -0.033 | -0.5848 | No | ||
| 64 | TYSND1 | 20007 | 15548 | -0.033 | -0.5830 | No | ||
| 65 | BCL6B | 20373 | 15551 | -0.033 | -0.5800 | No | ||
| 66 | ARF1 | 64 | 15748 | -0.038 | -0.5870 | No | ||
| 67 | AQP6 | 22364 8621 | 16324 | -0.061 | -0.6124 | No | ||
| 68 | SSH2 | 6202 | 16461 | -0.067 | -0.6136 | No | ||
| 69 | RPS4X | 9756 | 17106 | -0.113 | -0.6379 | No | ||
| 70 | NXN | 5204 | 17300 | -0.134 | -0.6359 | No | ||
| 71 | EXOSC2 | 15044 | 17684 | -0.178 | -0.6401 | Yes | ||
| 72 | PLXNA2 | 5269 | 17782 | -0.192 | -0.6276 | Yes | ||
| 73 | MTX1 | 15282 1918 | 18512 | -1.111 | -0.5642 | Yes | ||
| 74 | DGKA | 3359 19589 | 18527 | -1.227 | -0.4516 | Yes | ||
| 75 | RPL38 | 12562 20606 7475 | 18590 | -2.072 | -0.2634 | Yes | ||
| 76 | GAS7 | 20836 1299 | 18604 | -2.865 | 0.0006 | Yes |

