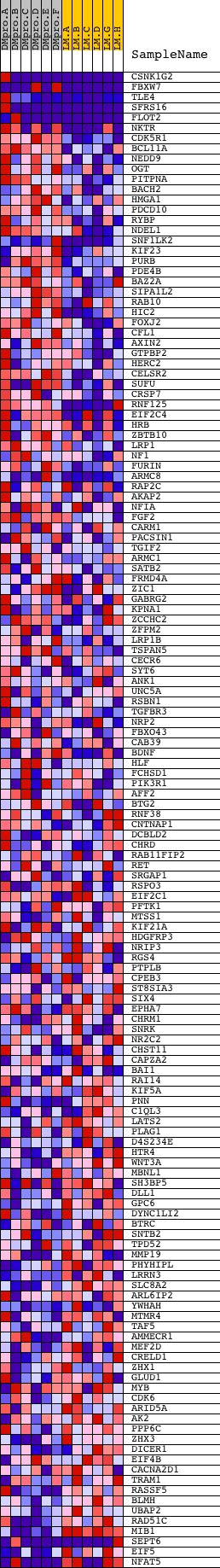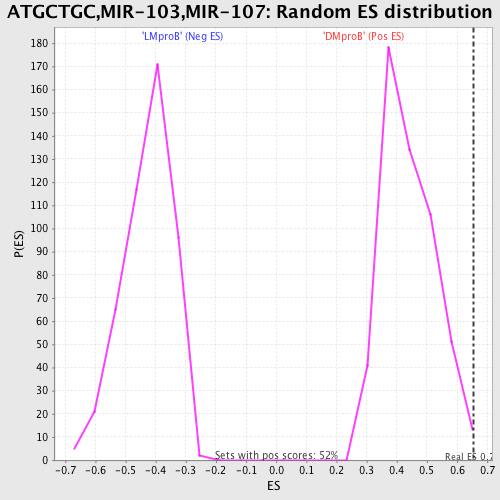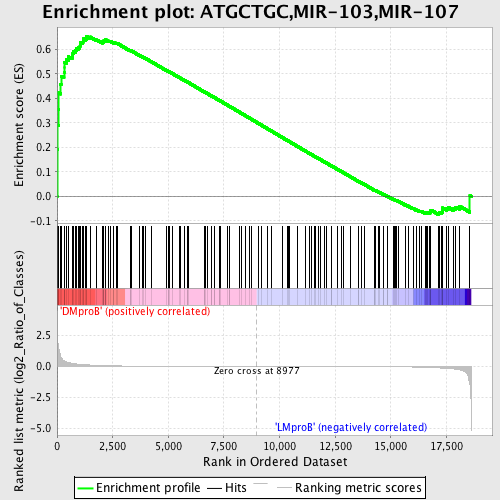
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | ATGCTGC,MIR-103,MIR-107 |
| Enrichment Score (ES) | 0.65383357 |
| Normalized Enrichment Score (NES) | 1.4874351 |
| Nominal p-value | 0.0076481835 |
| FDR q-value | 0.64155614 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CSNK1G2 | 19940 3418 | 5 | 4.027 | 0.1926 | Yes | ||
| 2 | FBXW7 | 1805 11928 | 33 | 2.037 | 0.2887 | Yes | ||
| 3 | TLE4 | 23720 3697 | 64 | 1.444 | 0.3562 | Yes | ||
| 4 | SFRS16 | 17943 3840 | 74 | 1.383 | 0.4220 | Yes | ||
| 5 | FLOT2 | 1247 8975 1430 | 159 | 0.843 | 0.4578 | Yes | ||
| 6 | NKTR | 9464 5175 | 182 | 0.712 | 0.4907 | Yes | ||
| 7 | CDK5R1 | 20745 | 310 | 0.455 | 0.5056 | Yes | ||
| 8 | BCL11A | 4691 | 318 | 0.446 | 0.5266 | Yes | ||
| 9 | NEDD9 | 3206 21483 | 320 | 0.441 | 0.5477 | Yes | ||
| 10 | OGT | 4241 24274 | 412 | 0.364 | 0.5602 | Yes | ||
| 11 | PITPNA | 20778 | 504 | 0.312 | 0.5702 | Yes | ||
| 12 | BACH2 | 8648 | 676 | 0.247 | 0.5727 | Yes | ||
| 13 | HMGA1 | 23323 | 705 | 0.236 | 0.5825 | Yes | ||
| 14 | PDCD10 | 15314 | 732 | 0.229 | 0.5920 | Yes | ||
| 15 | RYBP | 17056 | 829 | 0.206 | 0.5967 | Yes | ||
| 16 | NDEL1 | 20399 | 874 | 0.197 | 0.6037 | Yes | ||
| 17 | SNF1LK2 | 6168 10649 | 947 | 0.183 | 0.6086 | Yes | ||
| 18 | KIF23 | 19091 | 1025 | 0.171 | 0.6127 | Yes | ||
| 19 | PURB | 5335 | 1033 | 0.170 | 0.6204 | Yes | ||
| 20 | PDE4B | 9543 | 1057 | 0.166 | 0.6271 | Yes | ||
| 21 | BAZ2A | 4371 | 1144 | 0.154 | 0.6298 | Yes | ||
| 22 | SIPA1L2 | 10863 | 1166 | 0.150 | 0.6359 | Yes | ||
| 23 | RAB10 | 2090 21119 | 1176 | 0.149 | 0.6425 | Yes | ||
| 24 | HIC2 | 7185 | 1283 | 0.135 | 0.6432 | Yes | ||
| 25 | FOXJ2 | 17294 | 1319 | 0.130 | 0.6476 | Yes | ||
| 26 | CFL1 | 4516 | 1320 | 0.130 | 0.6538 | Yes | ||
| 27 | AXIN2 | 20618 | 1485 | 0.112 | 0.6503 | No | ||
| 28 | GTPBP2 | 23221 | 1784 | 0.087 | 0.6383 | No | ||
| 29 | HERC2 | 9085 | 2025 | 0.071 | 0.6287 | No | ||
| 30 | CELSR2 | 15194 12000 | 2069 | 0.067 | 0.6296 | No | ||
| 31 | SUFU | 6283 | 2078 | 0.067 | 0.6324 | No | ||
| 32 | CRSP7 | 18838 | 2104 | 0.065 | 0.6342 | No | ||
| 33 | RNF125 | 12559 23614 | 2167 | 0.062 | 0.6338 | No | ||
| 34 | EIF2C4 | 8043 13362 | 2177 | 0.061 | 0.6362 | No | ||
| 35 | HRB | 14208 4066 | 2185 | 0.061 | 0.6388 | No | ||
| 36 | ZBTB10 | 10468 | 2323 | 0.055 | 0.6340 | No | ||
| 37 | LRP1 | 9284 | 2391 | 0.052 | 0.6328 | No | ||
| 38 | NF1 | 5165 | 2532 | 0.046 | 0.6274 | No | ||
| 39 | FURIN | 9538 | 2548 | 0.045 | 0.6288 | No | ||
| 40 | ARMC8 | 7893 3056 13167 3144 | 2653 | 0.041 | 0.6252 | No | ||
| 41 | RAP2C | 24156 | 2693 | 0.040 | 0.6250 | No | ||
| 42 | AKAP2 | 8565 | 3283 | 0.026 | 0.5943 | No | ||
| 43 | NFIA | 16172 5170 | 3303 | 0.026 | 0.5945 | No | ||
| 44 | FGF2 | 15608 | 3330 | 0.025 | 0.5943 | No | ||
| 45 | CARM1 | 19539 | 3719 | 0.019 | 0.5742 | No | ||
| 46 | PACSIN1 | 6257 23322 10744 | 3831 | 0.018 | 0.5691 | No | ||
| 47 | TGIF2 | 6013 6012 | 3881 | 0.017 | 0.5672 | No | ||
| 48 | ARMC1 | 7907 | 3991 | 0.016 | 0.5621 | No | ||
| 49 | SATB2 | 5604 | 4247 | 0.014 | 0.5490 | No | ||
| 50 | FRMD4A | 5557 | 4938 | 0.010 | 0.5121 | No | ||
| 51 | ZIC1 | 19040 | 4985 | 0.010 | 0.5101 | No | ||
| 52 | GABRG2 | 1259 4748 8996 1331 | 5002 | 0.010 | 0.5097 | No | ||
| 53 | KPNA1 | 22776 | 5034 | 0.010 | 0.5085 | No | ||
| 54 | ZCCHC2 | 10409 | 5198 | 0.009 | 0.5001 | No | ||
| 55 | ZFPM2 | 22483 | 5490 | 0.008 | 0.4847 | No | ||
| 56 | LRP1B | 8249 | 5531 | 0.008 | 0.4829 | No | ||
| 57 | TSPAN5 | 15402 | 5705 | 0.007 | 0.4739 | No | ||
| 58 | CECR6 | 17018 | 5841 | 0.007 | 0.4669 | No | ||
| 59 | SYT6 | 13437 7042 12040 | 5859 | 0.007 | 0.4663 | No | ||
| 60 | ANK1 | 22501 18650 4385 8590 18649 | 5923 | 0.006 | 0.4632 | No | ||
| 61 | UNC5A | 3288 21631 | 6614 | 0.005 | 0.4260 | No | ||
| 62 | RSBN1 | 6035 | 6618 | 0.005 | 0.4261 | No | ||
| 63 | TGFBR3 | 16453 | 6684 | 0.004 | 0.4228 | No | ||
| 64 | NRP2 | 5194 9486 5195 | 6758 | 0.004 | 0.4190 | No | ||
| 65 | FBXO43 | 8123 | 6923 | 0.004 | 0.4103 | No | ||
| 66 | CAB39 | 8672 | 6942 | 0.004 | 0.4096 | No | ||
| 67 | BDNF | 14926 2797 | 7067 | 0.004 | 0.4030 | No | ||
| 68 | HLF | 10124 | 7090 | 0.004 | 0.4020 | No | ||
| 69 | FCHSD1 | 6661 11435 | 7281 | 0.003 | 0.3919 | No | ||
| 70 | PIK3R1 | 3170 | 7286 | 0.003 | 0.3918 | No | ||
| 71 | AFF2 | 8977 | 7322 | 0.003 | 0.3900 | No | ||
| 72 | BTG2 | 13832 | 7656 | 0.002 | 0.3721 | No | ||
| 73 | RNF38 | 7862 2328 13115 | 7735 | 0.002 | 0.3680 | No | ||
| 74 | CNTNAP1 | 11987 | 7767 | 0.002 | 0.3664 | No | ||
| 75 | DCBLD2 | 7855 | 8190 | 0.001 | 0.3437 | No | ||
| 76 | CHRD | 22817 | 8267 | 0.001 | 0.3396 | No | ||
| 77 | RAB11FIP2 | 23635 | 8464 | 0.001 | 0.3290 | No | ||
| 78 | RET | 17028 | 8625 | 0.001 | 0.3204 | No | ||
| 79 | SRGAP1 | 19615 | 8733 | 0.000 | 0.3146 | No | ||
| 80 | RSPO3 | 473 19797 | 9060 | -0.000 | 0.2970 | No | ||
| 81 | EIF2C1 | 10672 | 9174 | -0.000 | 0.2909 | No | ||
| 82 | PFTK1 | 16924 | 9460 | -0.001 | 0.2755 | No | ||
| 83 | MTSS1 | 5585 22285 | 9627 | -0.001 | 0.2666 | No | ||
| 84 | KIF21A | 4952 | 10137 | -0.002 | 0.2391 | No | ||
| 85 | HDGFRP3 | 6621 | 10334 | -0.002 | 0.2286 | No | ||
| 86 | NRIP3 | 17679 | 10410 | -0.002 | 0.2246 | No | ||
| 87 | RGS4 | 9724 | 10425 | -0.002 | 0.2240 | No | ||
| 88 | PTPLB | 22273 | 10796 | -0.003 | 0.2041 | No | ||
| 89 | CPEB3 | 5539 | 11179 | -0.004 | 0.1836 | No | ||
| 90 | ST8SIA3 | 23535 | 11334 | -0.004 | 0.1755 | No | ||
| 91 | SIX4 | 5445 | 11445 | -0.004 | 0.1697 | No | ||
| 92 | EPHA7 | 8909 4674 | 11590 | -0.005 | 0.1622 | No | ||
| 93 | CHRM1 | 23943 | 11616 | -0.005 | 0.1610 | No | ||
| 94 | SNRK | 5467 5468 9840 | 11738 | -0.005 | 0.1547 | No | ||
| 95 | NR2C2 | 10216 | 11854 | -0.005 | 0.1488 | No | ||
| 96 | CHST11 | 19921 | 12006 | -0.006 | 0.1409 | No | ||
| 97 | CAPZA2 | 8688 991 | 12123 | -0.006 | 0.1349 | No | ||
| 98 | BAI1 | 22459 | 12324 | -0.007 | 0.1244 | No | ||
| 99 | RAI14 | 22327 2191 | 12330 | -0.007 | 0.1244 | No | ||
| 100 | KIF5A | 9222 | 12607 | -0.007 | 0.1098 | No | ||
| 101 | PNN | 9596 | 12767 | -0.008 | 0.1016 | No | ||
| 102 | C1QL3 | 14684 | 12890 | -0.008 | 0.0954 | No | ||
| 103 | LATS2 | 11923 6928 | 13191 | -0.009 | 0.0796 | No | ||
| 104 | PLAG1 | 7148 15949 12161 | 13525 | -0.011 | 0.0620 | No | ||
| 105 | D4S234E | 16546 | 13533 | -0.011 | 0.0622 | No | ||
| 106 | HTR4 | 1987 4887 | 13660 | -0.012 | 0.0559 | No | ||
| 107 | WNT3A | 20431 | 13837 | -0.013 | 0.0470 | No | ||
| 108 | MBNL1 | 1921 15582 | 14268 | -0.015 | 0.0245 | No | ||
| 109 | SH3BP5 | 6278 | 14273 | -0.015 | 0.0250 | No | ||
| 110 | DLL1 | 23119 | 14298 | -0.016 | 0.0244 | No | ||
| 111 | GPC6 | 21937 | 14445 | -0.017 | 0.0173 | No | ||
| 112 | DYNC1LI2 | 331 | 14470 | -0.017 | 0.0169 | No | ||
| 113 | BTRC | 4459 | 14663 | -0.019 | 0.0074 | No | ||
| 114 | SNTB2 | 3887 18476 | 14691 | -0.019 | 0.0068 | No | ||
| 115 | TPD52 | 18830 5794 | 14854 | -0.021 | -0.0010 | No | ||
| 116 | MMP19 | 7191 | 14867 | -0.021 | -0.0006 | No | ||
| 117 | PHYHIPL | 12907 7711 | 15099 | -0.024 | -0.0119 | No | ||
| 118 | LRRN3 | 21077 | 15150 | -0.025 | -0.0134 | No | ||
| 119 | SLC8A2 | 18384 | 15211 | -0.026 | -0.0154 | No | ||
| 120 | ARL6IP2 | 1496 7090 12093 | 15257 | -0.027 | -0.0166 | No | ||
| 121 | YWHAH | 5937 10368 | 15332 | -0.028 | -0.0192 | No | ||
| 122 | MTMR4 | 20712 | 15646 | -0.036 | -0.0345 | No | ||
| 123 | TAF5 | 23833 5934 3759 | 15775 | -0.039 | -0.0395 | No | ||
| 124 | AMMECR1 | 7067 | 15996 | -0.047 | -0.0492 | No | ||
| 125 | MEF2D | 9379 | 16164 | -0.054 | -0.0557 | No | ||
| 126 | CRELD1 | 17329 | 16299 | -0.060 | -0.0601 | No | ||
| 127 | ZHX1 | 10428 5990 | 16368 | -0.063 | -0.0607 | No | ||
| 128 | GLUD1 | 9022 | 16549 | -0.072 | -0.0670 | No | ||
| 129 | MYB | 3344 3355 19806 | 16614 | -0.076 | -0.0668 | No | ||
| 130 | CDK6 | 16600 | 16668 | -0.079 | -0.0659 | No | ||
| 131 | ARID5A | 5668 | 16741 | -0.085 | -0.0657 | No | ||
| 132 | AK2 | 8563 2479 16073 | 16788 | -0.089 | -0.0640 | No | ||
| 133 | PPP6C | 7491 | 16789 | -0.089 | -0.0597 | No | ||
| 134 | ZHX3 | 6800 | 16803 | -0.090 | -0.0561 | No | ||
| 135 | DICER1 | 20989 | 17140 | -0.116 | -0.0687 | No | ||
| 136 | EIF4B | 13279 7979 | 17172 | -0.119 | -0.0647 | No | ||
| 137 | CACNA2D1 | 8676 | 17286 | -0.132 | -0.0645 | No | ||
| 138 | TRAM1 | 13016 14000 | 17315 | -0.135 | -0.0595 | No | ||
| 139 | RASSF5 | 13837 | 17324 | -0.136 | -0.0535 | No | ||
| 140 | BLMH | 20769 | 17337 | -0.138 | -0.0475 | No | ||
| 141 | UBAP2 | 15912 | 17506 | -0.155 | -0.0492 | No | ||
| 142 | RAD51C | 4330 | 17575 | -0.164 | -0.0450 | No | ||
| 143 | MIB1 | 5908 | 17803 | -0.195 | -0.0480 | No | ||
| 144 | SEPT6 | 2591 7132 | 17918 | -0.218 | -0.0437 | No | ||
| 145 | EIF5 | 5736 | 18081 | -0.267 | -0.0397 | No | ||
| 146 | NFAT5 | 3921 7037 12036 | 18556 | -1.432 | 0.0032 | No |